Download files from repositories
Several institutions host different kind of genomics data.
For example the genome browser Ensembl is also a public repository of genomes and annotation that can be freely downloaded and used for any kind of analysis.
The resource Ensembl Bacteria contains a large number of bacterial genomes and their annotation. As an example we can browse the page corresponding to Escherichia coli ‘BL21-Gold(DE3)pLysS AG’

We can click on “Download genes, cDNAs, ncRNA, proteins FASTA”

And then on DNA
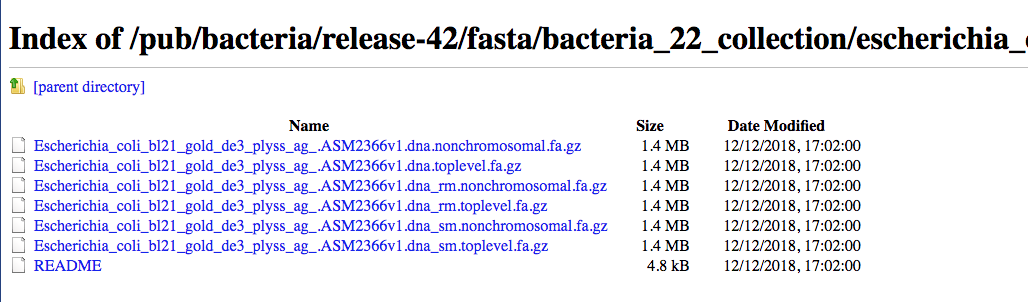
Then as an example we can use the copy the link address of the README file using the mouse right button.
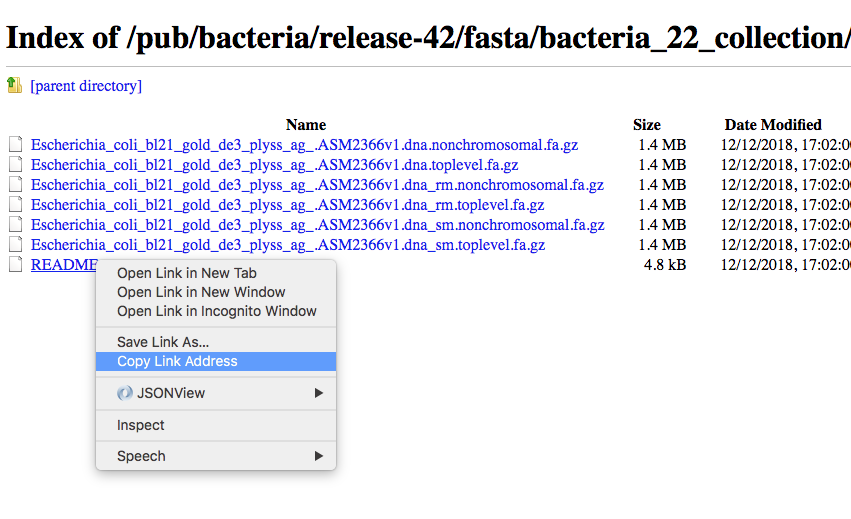
Then we can go back to our command line and use the program wget to download that file and using CTRL+C to paste the address:
wget ftp://ftp.ensemblgenomes.org/pub/bacteria/release-42/fasta/bacteria_22_collection/escherichia_coli_bl21_gold_de3_plyss_ag_/dna/README
--2019-03-06 18:59:13-- ftp://ftp.ensemblgenomes.org/pub/bacteria/release-42/fasta/bacteria_22_collection/escherichia_coli_bl21_gold_de3_plyss_ag_/dna/README
=> ‘README’
Resolving ftp.ensemblgenomes.org (ftp.ensemblgenomes.org)... 193.62.197.94
Connecting to ftp.ensemblgenomes.org (ftp.ensemblgenomes.org)|193.62.197.94|:21... connected.
Logging in as anonymous ... Logged in!
==> SYST ... done. ==> PWD ... done.
==> TYPE I ... done. ==> CWD (1) /pub/bacteria/release-42/fasta/bacteria_22_collection/escherichia_coli_bl21_gold_de3_plyss_ag_/dna ... done.
==> SIZE README ... 4923
==> PASV ... done. ==> RETR README ... done.
Length: 4923 (4.8K) (unauthoritative)
100%[======================================================================================================================>] 4,923 --.-K/s in 0s
2019-03-06 18:59:14 (295 MB/s) - ‘README’ saved [4923]
we can then use the program more to display part of the content of the file:
more README
#### README ####
IMPORTANT: Please note you can download correlation data tables,
supported by Ensembl, via the highly customisable BioMart and
EnsMart data mining tools. See http://www.ensembl.org/biomart/martview or
http://www.ebi.ac.uk/biomart/ for more information.
The genome assembly represented here corresponds to
GCA_000023665.1
#######################
Fasta DNA dumps
#######################
-----------
FILE NAMES
------------
The files are consistently named following this pattern:
<species>.<assembly>.<sequence type>.<id type>.<id>.fa.gz
<species>: The systematic name of the species.
<assembly>: The assembly build name.
<sequence type>:
* 'dna' - unmasked genomic DNA sequences.
--More--(14%)
Pressing the bar allows us to scroll down the file, while for exiting you just click CTRL+C.
After reading the README we can download the file named toplevel that contains chromosomes, regions not assembled into chromosomes and N padded haplotype/patch regions:
wget ftp://ftp.ensemblgenomes.org/pub/bacteria/release-42/fasta/bacteria_22_collection/escherichia_coli_bl21_gold_de3_plyss_ag_/dna/Escherichia_coli_bl21_gold_de3_plyss_ag_.ASM2366v1.dna.toplevel.fa.gz
We can use the options -lh of the program ls to list attributes of the files and show in human readable format the size fo the files
ls -lh
total 2.0M
drwxr-xr-x 5 lcozzuto Bioinformatics_Unit 209 Mar 7 11:48 advanced_linux_2019
-rw-r--r-- 1 lcozzuto Bioinformatics_Unit 1.4M Mar 7 13:06 Escherichia_coli_bl21_gold_de3_plyss_ag_.ASM2366v1.dna.toplevel.fa.gz
drwxr-xr-x 2 lcozzuto Bioinformatics_Unit 39 Mar 6 18:17 my_beautiful_folder
-rw-r--r-- 1 lcozzuto Bioinformatics_Unit 0 Mar 6 18:15 my_ugly_file.txt
drwxr-xr-x 2 lcozzuto Bioinformatics_Unit 34 Mar 6 18:17 my_ugly_folder
-rw-r--r-- 1 lcozzuto Bioinformatics_Unit 4.9K Mar 6 18:59 README
For unzipping the file we can use the program gunzip.
The uncompressed file is now 4.5M.
Let’s see the content of the file.
more Escherichia_coli_bl21_gold_de3_plyss_ag_.ASM2366v1.dna.toplevel.fa
>CP001665 dna:supercontig supercontig:ASM2366v1:CP001665:1:4570938:1 REF
CGTCCTGGATCTTTATTAGATCGATTAAGCCAATTTTTGTCTATGGTCATTAAATTTTCC
AATATGCGGCGTAAATCGTGCCCGCCTCGCGGCAGGATCGTTTACACTTAGCGAGTTCTG
GAAAGTCCTGTGGATAAATCGGGAAAATCTGTGAGAAACAGAAGATCTCTTGCGCAGTTT
AGGCTATGATCCGCGGTCCCGATCGTTTTGCAGGATCTTGATCGGGCATATAACCGCAGA
CAGCGGTTCGTGCGTCACCCTCAAGCAGGGTCTTTTCGACGTACGTCAACAATCATGAAT
GTTTCAGCCTTAGTCATTATCGACTTTTGTTCGAGTGGAGTCCGCCGTGTCACTTTCGCT
TTGGCAGCAGTGTCTTGCCCGATTGCAGGATGAGTTACCAGCCACAGAATTCAGTATGTG
GATACGCCCATTGCAGGCGGAACTGAGCGATAACACGCTGGCCCTGTACGCGCCAAACCG
TTTTGTCCTCGATTGGGTACGGGACAAGTACCTTAATAATATCAATGGACTGCTAACCAG
TTTCTGCGGAGCGGATGCCCCACAGCTGCGTTTTGAAGTCGGCACCAAACCGGTGACGCA
AACGCCACAAGCGGCAGTGACGAGCAACGTCGCGGCCCCTGCACAGGTGGCGCAAACGCA
GCCGCAACGTGCTGCGCCTTCTACGCGCTCAGGTTGGGATAACGTCCCGGCCCCGGCAGA
ACCGACCTATCGTTCTAACGTAAACGTCAAACACACGTTTGATAACTTCGTTGAAGGTAA
ATCTAACCAACTGGCGCGCGCGGCGGCTCGCCAGGTGGCGGATAACCCTGGCGGTGCCTA
TAACCCGTTGTTCCTTTATGGCGGCACGGGTCTGGGTAAAACTCACCTGCTGCATGCGGT
GGGTAACGGCATTATGGCGCGCAAGCCGAATGCCAAAGTGGTTTATATGCACTCCGAGCG
CTTTGTTCAGGACATGGTTAAAGCCCTGCAAAACAACGCGATCGAAGAGTTTAAACGCTA
CTACCGTTCCGTAGATGCACTGCTGATCGACGATATTCAGTTTTTTGCTAATAAAGAACG
ATCTCAGGAAGAGTTTTTCCACACCTTCAACGCCCTGCTGGAAGGTAATCAACAGATCAT
TCTCACCTCGGATCGCTATCCGAAAGAGATCAACGGCGTTGAGGATCGTTTGAAATCCCG
CTTCGGTTGGGGACTGACTGTGGCGATCGAACCGCCAGAGCTGGAAACCCGTGTGGCGAT
CCTGATGAAAAAGGCCGACGAAAACGACATTCGTTTGCCGGGCGAAGTGGCGTTCTTTAT
CGCCAAGCGTCTACGATCTAACGTACGTGAGCTGGAAGGGGCGCTGAACCGCGTCATTGC
The file contains the whole genome of the bacteria.
The first line contains the character > and the name of the molecule / genome.
This format is called FASTA format and is universally used for storing one or multiple DNA/RNA/Protein sequences.
We can now download in the same ways the proteins:

and after unzipping the file we can have a look at it.
more Escherichia_coli_bl21_gold_de3_plyss_ag_.ASM2366v1.pep.all.fa
>ACT27082 pep supercontig:ASM2366v1:CP001665:347:1750:1 gene:ECBD_0001 transcript:ACT27082 gene_biotype:protein_coding transcript_biotype:protein_coding descrip
tion:chromosomal replication initiator protein DnaA
MSLSLWQQCLARLQDELPATEFSMWIRPLQAELSDNTLALYAPNRFVLDWVRDKYLNNIN
GLLTSFCGADAPQLRFEVGTKPVTQTPQAAVTSNVAAPAQVAQTQPQRAAPSTRSGWDNV
PAPAEPTYRSNVNVKHTFDNFVEGKSNQLARAAARQVADNPGGAYNPLFLYGGTGLGKTH
LLHAVGNGIMARKPNAKVVYMHSERFVQDMVKALQNNAIEEFKRYYRSVDALLIDDIQFF
ANKERSQEEFFHTFNALLEGNQQIILTSDRYPKEINGVEDRLKSRFGWGLTVAIEPPELE
TRVAILMKKADENDIRLPGEVAFFIAKRLRSNVRELEGALNRVIANANFTGRAITIDFVR
EALRDLLALQEKLVTIDNIQKTVAEYYKIKVADLLSKRRSRSVARPRQMAMALAKELTNH
SLPEIGDAFGGRDHTTVLHACRKIEQLREESHDIKEDFSNLIRTLSS
>ACT27083 pep supercontig:ASM2366v1:CP001665:1755:2855:1 gene:ECBD_0002 transcript:ACT27083 gene_biotype:protein_coding transcript_biotype:protein_coding descri
ption:DNA polymerase III, beta subunit
MKFTVEREHLLKPLQQVSGPLGGRPTLPILGNLLLQVADGTLSLTGTDLEMEMVARVALV
QPHEPGATTVPARKFFDICRGLPEGAEIAVQLEGERMLVRSGRSRFSLSTLPAADFPNLD
DWQSEVEFTLPQATMKRLIEATQFSMAHQDVRYYLNGMLFETEGEELRTVATDGHRLAVC
SMPIGQSLPSHSVIVPRKGVIELMRMLDGGDNPLRVQIGSNNIRAHVGDFIFTSKLVDGR
FPDYRRVLPKNPDKHLEAGCDLLKQAFARAAILSNEKFRGVRLYVSENQLKITANNPEQE
EAEEILDVTYSGAEMEIGFNVSYVLDVLNALKCENVRMMLTDSVSSVQIEDAASQSAAYV
VMPMRL
>ACT27084 pep supercontig:ASM2366v1:CP001665:2855:3928:1 gene:ECBD_0003 transcript:ACT27084 gene_biotype:protein_coding transcript_biotype:protein_coding descri
ption:DNA replication and repair protein RecF
MSLTRLLIRDFRNIETADLALSPGFNFLVGANGSGKTSVLEAIYTLGHGRAFRSLQIGRV
IRHEQEAFVLHGRLQGEERETAIGLTKDKQGDSKVRIDGTDGHKVAELAHLMPMQLITPE
GFTLLNGGPKYRRAFLDWGCFHNEPGFFTAWSNLKRLLKQRNAALRQVTRYEQLRPWDKE
--More--(0%)
We see that many protein sequences are embedded in the files and separated by their name, always preceded by the character ”>”.
To know how many sequences are in the files we can use the program grep with the option -c for counting the number of rows containg the character ”>”:
grep ">" -c Escherichia_coli_bl21_gold_de3_plyss_ag_.ASM2366v1.pep.all.fa
4228
Recap
- wget downloads file from a URL
- more prints a part of the content of a file in interactive way
- grep extract the rows containing a particular character / pattern.