NanoMod
This module allows to predict the loci with RNA modifications starting from data produced by NanoPreprocess.
Workflow
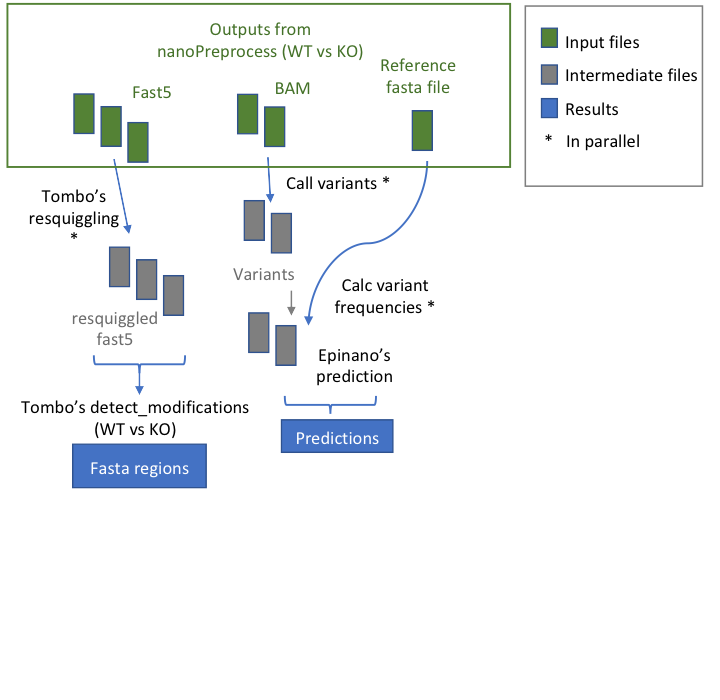
- index_reference index the reference file for Epinano
- call_variants uses Samtools for calling the variants for Epinano
- calc_var_frequencies it uses TSV_to_Variants_Freq.py3 for calculating the frequencies of each variants for Epinano
- predict_with_EPInano It predicts the modifications with Epinano in parallel splitting the input file in 1 million rows
- combineEpinanoPred It combine the results from Epinano
- resquiggling resquiggle fast5 files for Tombo
- getModifications it estimates the modifications using Tombo comparing WT vs KO
Input Parameters
- input_path path to the folders produced by NanoPreprocessing step.
- comparison tab separated text file containing the list of comparison. Here an example:
WT1 KO1
WT2 KO2
WT3 KO3
- reference reference transcriptome
- output folder
- coverage read coverage threshold for prediction
- tombo_opt options for tombo
- epinano_opt options for epinano
Results
Three folders are produced by this module:
- Epinano, containing the results obtained with this method. You have a single file with putative modifications:
#Kmer,Window,Ref,Coverage,q1,q2,q3,q4,q5,mis1,mis2,mis3,mis4,mis5,ins1,ins2,ins3,ins4,ins5,del1,del2,del3,del4,del5,prediction,dist,ProbM,Pro
bU
AGTGG,394404:394405:394406:394407:394408,chr2,8.0:8.0:7.0:7.0:7.0,21.5,21.25,19.857,23.0,16.285999999999998,0.0,0.0,0.0,0.0,0.0,0.0,0.062,0.0
71,0.0,0.0,0.0,0.0,0.0,0.0,0.0,unm,19.26143361547619,3.00000089999998e-14,0.9999999999999699
TTTTT,12150:12151:12152:12153:12154,chr8,3.0:3.0:3.0:3.0:3.0,0.0,16.5,18.5,16.0,16.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,0.0,1.0,0.3329999999
9999996,0.33299999999999996,0.0,0.0,unm,2.5976484688977424,0.06071658133381308,0.9392834186661868
ACATT,438165:438166:438167:438168:438169,chr13,67.0:67.0:67.0:68.0:68.0,13.635,13.446,9.323,9.6,12.127,0.03,0.045,0.015,0.147,0.0740000000000
0001,0.0,0.0,0.0,0.0,0.0,0.06,0.03,0.075,0.11800000000000001,0.07400000000000001,unm,0.08435556637195174,0.519879422458087,0.4801205775419129
5...
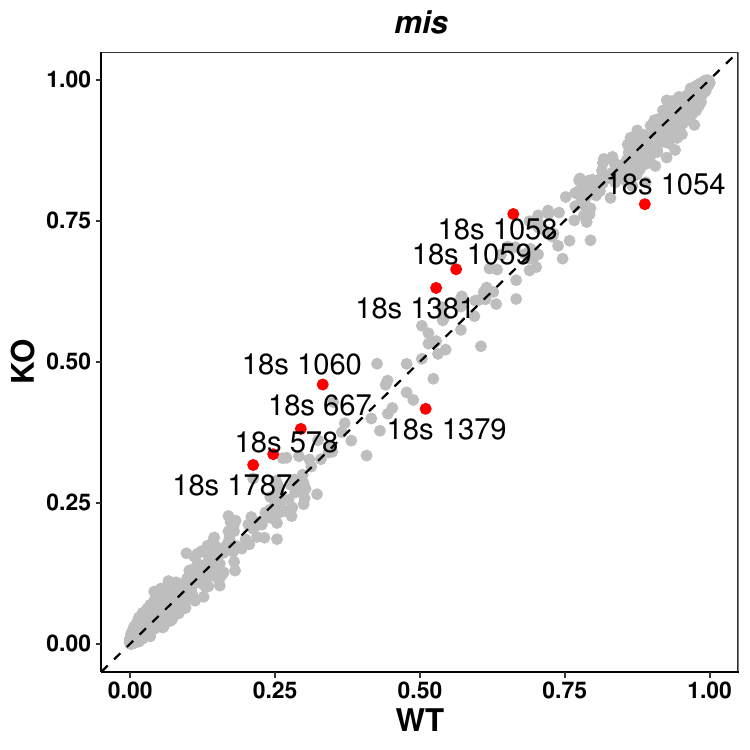
and three plots in pdf indicating possible events related to insertion, deletion and mismatches, see the example below.
- Tombo, containing the results obtained with this method in fasta format. You have one file for each comparison WT vs KO
>chr11:455562:- Est. Frac. Alternate: 0.98
TGACA
>chr12:1008723:- Est. Frac. Alternate: 0.98
TATCT
>chr15:491587:+ Est. Frac. Alternate: 0.96
TATAT
>chr10:425794:- Est. Frac. Alternate: 0.95
ATGTT
>chr13:510759:+ Est. Frac. Alternate: 0.95
...
And for convenience a 6 bed files with the coordinates of the event.

