Nextflow 3
Adding more processes
We can build a pipeline incrementally adding more and more processes. Nextflow will take care of the dependencies between the input / output and of the parallelization.
Let’s add to the test2.nf pipeline two additional steps, indexing of the reference genome and the read alignment using Bowtie. For that we will have to modify the test2.nf, params.config and nexflow.config files (the new script is available in the test3 folder).
In params.config, we have to add new parameters:
params {
reads = "$baseDir/../../testdata/*.fastq.gz"
reference = "$baseDir/../../testdata/chr19.fasta.gz"
outdir = "$baseDir"
//outdir = "s3://class-bucket-1/results"
email = "myemail@google.com"
}
In test3.nf, we have to add a new input for the reference sequence:
#!/usr/bin/env nextflow
/*
* This code enables the new dsl of Nextflow.
*/
nextflow.enable.dsl=2
/*
* NextFlow test pipe
* @authors
* Luca Cozzuto <lucacozzuto@gmail.com>
*
*/
/*
* Input parameters: read pairs
* Params are stored in the params.config file
*/
version = "1.0"
// this prevents a warning of undefined parameter
params.help = false
// this prints the input parameters
log.info """
BIOCORE@CRG - N F TESTPIPE ~ version ${version}
=============================================
reads : ${params.reads}
reference : ${params.reference}
outdir : ${params.outdir}
"""
// this prints the help in case you use --help parameter in the command line and it stops the pipeline
if (params.help) {
log.info 'This is the Biocore\'s NF test pipeline'
log.info 'Enjoy!'
log.info '\n'
exit 1
}
/*
* Defining the output folders.
*/
fastqcOutputFolder = "${params.outdir}/ouptut_fastqc"
alnOutputFolder = "${params.outdir}/ouptut_aln"
multiqcOutputFolder = "${params.outdir}/ouptut_multiQC"
/* Reading the file list and creating a "Channel": a queue that connects different channels.
* The queue is consumed by channels, so you cannot re-use a channel for different processes.
* If you need the same data for different processes you need to make more channels.
*/
Channel
.fromPath( params.reads )
.ifEmpty { error "Cannot find any reads matching: ${params.reads}" }
.set {reads}
reference = file(params.reference)
/*
* Process 1. Run FastQC on raw data. A process is the element for executing scripts / programs etc.
*/
process fastQC {
publishDir fastqcOutputFolder
tag { "${reads}" }
input:
path reads
output:
path "*_fastqc.*"
script:
"""
fastqc ${reads}
"""
}
/*
* Process 2. Bowtie index
*/
process bowtieIdx {
tag { "${ref}" }
input:
path ref
output:
tuple val("${ref}"), path ("${ref}*.ebwt")
script:
"""
gunzip -c ${ref} > reference.fa
bowtie-build reference.fa ${ref}
rm reference.fa
"""
}
/*
* Process 3. Bowtie alignment
*/
process bowtieAln {
publishDir alnOutputFolder, pattern: '*.sam'
tag { "${reads}" }
label 'twocpus'
input:
tuple val(refname), path (ref_files)
path reads
output:
path "${reads}.sam", emit: samples_sam
path "${reads}.log", emit: samples_log
script:
"""
bowtie -p ${task.cpus} ${refname} -q ${reads} -S > ${reads}.sam 2> ${reads}.log
"""
}
/*
/*
* Process 4. Run multiQC on fastQC results
*/
process multiQC {
publishDir multiqcOutputFolder, mode: 'copy' // this time do not link but copy the output file
input:
path (inputfiles)
output:
path("multiqc_report.html")
script:
"""
multiqc .
"""
}
workflow {
reads.view()
fastqc_out = fastQC(reads)
bowtie_index = bowtieIdx(reference)
bowtieAln(bowtie_index, reads)
multiQC(fastqc_out.mix(bowtieAln.out.samples_log).collect())
}
workflow.onComplete {
println ( workflow.success ? "\nDone! Open the following report in your browser --> ${multiqcOutputFolder}/multiqc_report.html\n" : "Oops .. something went wrong" )
}
This way, the singleton channel called reference is created. Its content can be used indefinitely. We also add a path specifying where to place the output files.
/*
* Defining the output folders.
*/
fastqcOutputFolder = "${params.outdir}/output_fastqc"
alnOutputFolder = "${params.outdir}/output_aln"
multiqcOutputFolder = "${params.outdir}/output_multiQC"
And we have to add two new processes. The first one is for the indexing the reference genome (with bowtie-build):
#!/usr/bin/env nextflow
/*
* This code enables the new dsl of Nextflow.
*/
nextflow.enable.dsl=2
/*
* NextFlow test pipe
* @authors
* Luca Cozzuto <lucacozzuto@gmail.com>
*
*/
/*
* Input parameters: read pairs
* Params are stored in the params.config file
*/
version = "1.0"
// this prevents a warning of undefined parameter
params.help = false
// this prints the input parameters
log.info """
BIOCORE@CRG - N F TESTPIPE ~ version ${version}
=============================================
reads : ${params.reads}
reference : ${params.reference}
outdir : ${params.outdir}
"""
// this prints the help in case you use --help parameter in the command line and it stops the pipeline
if (params.help) {
log.info 'This is the Biocore\'s NF test pipeline'
log.info 'Enjoy!'
log.info '\n'
exit 1
}
/*
* Defining the output folders.
*/
fastqcOutputFolder = "${params.outdir}/ouptut_fastqc"
alnOutputFolder = "${params.outdir}/ouptut_aln"
multiqcOutputFolder = "${params.outdir}/ouptut_multiQC"
/* Reading the file list and creating a "Channel": a queue that connects different channels.
* The queue is consumed by channels, so you cannot re-use a channel for different processes.
* If you need the same data for different processes you need to make more channels.
*/
Channel
.fromPath( params.reads )
.ifEmpty { error "Cannot find any reads matching: ${params.reads}" }
.set {reads}
reference = file(params.reference)
/*
* Process 1. Run FastQC on raw data. A process is the element for executing scripts / programs etc.
*/
process fastQC {
publishDir fastqcOutputFolder
tag { "${reads}" }
input:
path reads
output:
path "*_fastqc.*"
script:
"""
fastqc ${reads}
"""
}
/*
* Process 2. Bowtie index
*/
process bowtieIdx {
tag { "${ref}" }
input:
path ref
output:
tuple val("${ref}"), path ("${ref}*.ebwt")
script:
"""
gunzip -c ${ref} > reference.fa
bowtie-build reference.fa ${ref}
rm reference.fa
"""
}
/*
* Process 3. Bowtie alignment
*/
process bowtieAln {
publishDir alnOutputFolder, pattern: '*.sam'
tag { "${reads}" }
label 'twocpus'
input:
tuple val(refname), path (ref_files)
path reads
output:
path "${reads}.sam", emit: samples_sam
path "${reads}.log", emit: samples_log
script:
"""
bowtie -p ${task.cpus} ${refname} -q ${reads} -S > ${reads}.sam 2> ${reads}.log
"""
}
/*
/*
* Process 4. Run multiQC on fastQC results
*/
process multiQC {
publishDir multiqcOutputFolder, mode: 'copy' // this time do not link but copy the output file
input:
path (inputfiles)
output:
path("multiqc_report.html")
script:
"""
multiqc .
"""
}
workflow {
reads.view()
fastqc_out = fastQC(reads)
bowtie_index = bowtieIdx(reference)
bowtieAln(bowtie_index, reads)
multiQC(fastqc_out.mix(bowtieAln.out.samples_log).collect())
}
workflow.onComplete {
println ( workflow.success ? "\nDone! Open the following report in your browser --> ${multiqcOutputFolder}/multiqc_report.html\n" : "Oops .. something went wrong" )
}
Since bowtie indexing requires unzipped reference fasta file, we first gunzip it, then build the reference index, and finally remove the unzipped file.
The output channel is organized as a tuple; i.e., a list of elements.
The first element of the list is the name of the index as a value, the second is a list of files constituting the index.
The former is needed for building the command line of the alignment step, the latter are the files needed for the alignment.
The second process bowtieAln is the alignment step:
#!/usr/bin/env nextflow
/*
* This code enables the new dsl of Nextflow.
*/
nextflow.enable.dsl=2
/*
* NextFlow test pipe
* @authors
* Luca Cozzuto <lucacozzuto@gmail.com>
*
*/
/*
* Input parameters: read pairs
* Params are stored in the params.config file
*/
version = "1.0"
// this prevents a warning of undefined parameter
params.help = false
// this prints the input parameters
log.info """
BIOCORE@CRG - N F TESTPIPE ~ version ${version}
=============================================
reads : ${params.reads}
reference : ${params.reference}
outdir : ${params.outdir}
"""
// this prints the help in case you use --help parameter in the command line and it stops the pipeline
if (params.help) {
log.info 'This is the Biocore\'s NF test pipeline'
log.info 'Enjoy!'
log.info '\n'
exit 1
}
/*
* Defining the output folders.
*/
fastqcOutputFolder = "${params.outdir}/ouptut_fastqc"
alnOutputFolder = "${params.outdir}/ouptut_aln"
multiqcOutputFolder = "${params.outdir}/ouptut_multiQC"
/* Reading the file list and creating a "Channel": a queue that connects different channels.
* The queue is consumed by channels, so you cannot re-use a channel for different processes.
* If you need the same data for different processes you need to make more channels.
*/
Channel
.fromPath( params.reads )
.ifEmpty { error "Cannot find any reads matching: ${params.reads}" }
.set {reads}
reference = file(params.reference)
/*
* Process 1. Run FastQC on raw data. A process is the element for executing scripts / programs etc.
*/
process fastQC {
publishDir fastqcOutputFolder
tag { "${reads}" }
input:
path reads
output:
path "*_fastqc.*"
script:
"""
fastqc ${reads}
"""
}
/*
* Process 2. Bowtie index
*/
process bowtieIdx {
tag { "${ref}" }
input:
path ref
output:
tuple val("${ref}"), path ("${ref}*.ebwt")
script:
"""
gunzip -c ${ref} > reference.fa
bowtie-build reference.fa ${ref}
rm reference.fa
"""
}
/*
* Process 3. Bowtie alignment
*/
process bowtieAln {
publishDir alnOutputFolder, pattern: '*.sam'
tag { "${reads}" }
label 'twocpus'
input:
tuple val(refname), path (ref_files)
path reads
output:
path "${reads}.sam", emit: samples_sam
path "${reads}.log", emit: samples_log
script:
"""
bowtie -p ${task.cpus} ${refname} -q ${reads} -S > ${reads}.sam 2> ${reads}.log
"""
}
/*
/*
* Process 4. Run multiQC on fastQC results
*/
process multiQC {
publishDir multiqcOutputFolder, mode: 'copy' // this time do not link but copy the output file
input:
path (inputfiles)
output:
path("multiqc_report.html")
script:
"""
multiqc .
"""
}
workflow {
reads.view()
fastqc_out = fastQC(reads)
bowtie_index = bowtieIdx(reference)
bowtieAln(bowtie_index, reads)
multiQC(fastqc_out.mix(bowtieAln.out.samples_log).collect())
}
workflow.onComplete {
println ( workflow.success ? "\nDone! Open the following report in your browser --> ${multiqcOutputFolder}/multiqc_report.html\n" : "Oops .. something went wrong" )
}
There are two different input channels, the index and reads.
The index name specified by refname is used for building the command line; while the index files, indicated by ref_files, are linked to the current directory by using the path qualifier.
We also produced two kind of outputs, the alignments and logs. The first one is the one we want to keep as a final result; for that, we specify the pattern parameter in publishDir.
publishDir alnOutputFolder, pattern: '*.sam'
The second output will be passed to the next process, that is, the multiQC process. To distinguish the outputs let’s assign them different names.
output:
path "${reads}.sam", emit: samples_sam
path "${reads}.log", emit: samples_log
This section will allow us to connect these outputs directly with other processes when we call them in the workflow section:
#!/usr/bin/env nextflow
/*
* This code enables the new dsl of Nextflow.
*/
nextflow.enable.dsl=2
/*
* NextFlow test pipe
* @authors
* Luca Cozzuto <lucacozzuto@gmail.com>
*
*/
/*
* Input parameters: read pairs
* Params are stored in the params.config file
*/
version = "1.0"
// this prevents a warning of undefined parameter
params.help = false
// this prints the input parameters
log.info """
BIOCORE@CRG - N F TESTPIPE ~ version ${version}
=============================================
reads : ${params.reads}
reference : ${params.reference}
outdir : ${params.outdir}
"""
// this prints the help in case you use --help parameter in the command line and it stops the pipeline
if (params.help) {
log.info 'This is the Biocore\'s NF test pipeline'
log.info 'Enjoy!'
log.info '\n'
exit 1
}
/*
* Defining the output folders.
*/
fastqcOutputFolder = "${params.outdir}/ouptut_fastqc"
alnOutputFolder = "${params.outdir}/ouptut_aln"
multiqcOutputFolder = "${params.outdir}/ouptut_multiQC"
/* Reading the file list and creating a "Channel": a queue that connects different channels.
* The queue is consumed by channels, so you cannot re-use a channel for different processes.
* If you need the same data for different processes you need to make more channels.
*/
Channel
.fromPath( params.reads )
.ifEmpty { error "Cannot find any reads matching: ${params.reads}" }
.set {reads}
reference = file(params.reference)
/*
* Process 1. Run FastQC on raw data. A process is the element for executing scripts / programs etc.
*/
process fastQC {
publishDir fastqcOutputFolder
tag { "${reads}" }
input:
path reads
output:
path "*_fastqc.*"
script:
"""
fastqc ${reads}
"""
}
/*
* Process 2. Bowtie index
*/
process bowtieIdx {
tag { "${ref}" }
input:
path ref
output:
tuple val("${ref}"), path ("${ref}*.ebwt")
script:
"""
gunzip -c ${ref} > reference.fa
bowtie-build reference.fa ${ref}
rm reference.fa
"""
}
/*
* Process 3. Bowtie alignment
*/
process bowtieAln {
publishDir alnOutputFolder, pattern: '*.sam'
tag { "${reads}" }
label 'twocpus'
input:
tuple val(refname), path (ref_files)
path reads
output:
path "${reads}.sam", emit: samples_sam
path "${reads}.log", emit: samples_log
script:
"""
bowtie -p ${task.cpus} ${refname} -q ${reads} -S > ${reads}.sam 2> ${reads}.log
"""
}
/*
/*
* Process 4. Run multiQC on fastQC results
*/
process multiQC {
publishDir multiqcOutputFolder, mode: 'copy' // this time do not link but copy the output file
input:
path (inputfiles)
output:
path("multiqc_report.html")
script:
"""
multiqc .
"""
}
workflow {
reads.view()
fastqc_out = fastQC(reads)
bowtie_index = bowtieIdx(reference)
bowtieAln(bowtie_index, reads)
multiQC(fastqc_out.mix(bowtieAln.out.samples_log).collect())
}
workflow.onComplete {
println ( workflow.success ? "\nDone! Open the following report in your browser --> ${multiqcOutputFolder}/multiqc_report.html\n" : "Oops .. something went wrong" )
}
As you can see, we passed the samples_log output to the multiqc process after mixing it with the output channel from the fastqc process.
Profiles
For deploying a pipeline in a cluster or Cloud, in the nextflow.config file, we need to indicate what kind of the executor to use.
In the Nextflow framework architecture, the executor indicates which the batch-queue system to use to submit jobs to a HPC or to Cloud.
The executor is completely abstracted, so you can switch from SGE to SLURM just by changing this parameter in the configuration file.
You can group different classes of configuration or profiles within a single nextflow.config file.
Let’s inspect the nextflow.config file in test3 folder. We can see three different profiles:
standard
cluster
cloud
The first profile indicates the resources needed for running the pipeline locally. They are quite small since we have little power and CPU on the test node.
includeConfig "$baseDir/params.config"
process.container = 'biocorecrg/c4lwg-2018:latest'
singularity.cacheDir = "$baseDir/singularity"
profiles {
standard {
process {
containerOptions = { workflow.containerEngine == "docker" ? '-u $(id -u):$(id -g)': null}
executor="local"
memory='0.6G'
cpus='1'
time='6h'
withLabel: 'twocpus' {
memory='0.6G'
cpus='1'
}
}
}
cluster {
process {
containerOptions = { workflow.containerEngine == "docker" ? '-u $(id -u):$(id -g)': null}
executor="SGE"
queue = "smallcpus"
memory='1G'
cpus='1'
time='6h'
withLabel: 'twocpus' {
queue = "bigcpus"
memory='4G'
cpus='2'
}
}
}
cloud {
workDir = 's3://nf-class-bucket-XXX/work'
aws.region = 'eu-central-1'
aws.batch.cliPath = '/home/ec2-user/miniconda/bin/aws'
process {
executor = 'awsbatch'
queue = 'spot'
memory='1G'
cpus='1'
time='6h'
withLabel: 'twocpus' {
memory='2G'
cpus='2'
}
}
}
}
As you can see, we explicitly indicated the local executor. By definition, the local executor is a default executor if the pipeline is run without specifying a profile.
The second profile is for running the pipeline on the cluster; here in particular for the cluster supporting the Sun Grid Engine queuing system:
includeConfig "$baseDir/params.config"
process.container = 'biocorecrg/c4lwg-2018:latest'
singularity.cacheDir = "$baseDir/singularity"
profiles {
standard {
process {
containerOptions = { workflow.containerEngine == "docker" ? '-u $(id -u):$(id -g)': null}
executor="local"
memory='0.6G'
cpus='1'
time='6h'
withLabel: 'twocpus' {
memory='0.6G'
cpus='1'
}
}
}
cluster {
process {
containerOptions = { workflow.containerEngine == "docker" ? '-u $(id -u):$(id -g)': null}
executor="SGE"
queue = "smallcpus"
memory='1G'
cpus='1'
time='6h'
withLabel: 'twocpus' {
queue = "bigcpus"
memory='4G'
cpus='2'
}
}
}
cloud {
workDir = 's3://nf-class-bucket-XXX/work'
aws.region = 'eu-central-1'
aws.batch.cliPath = '/home/ec2-user/miniconda/bin/aws'
process {
executor = 'awsbatch'
queue = 'spot'
memory='1G'
cpus='1'
time='6h'
withLabel: 'twocpus' {
memory='2G'
cpus='2'
}
}
}
}
This profile indicates that the system uses Sun Grid Engine as a job scheduler and that we have different queues for small jobs and more intensive ones.
Deployment in the AWS cloud
The final profile is for running the pipeline in the Amazon Cloud, known as Amazon Web Services or AWS. In particular, we will use AWS Batch that allows the execution of containerised workloads in the Amazon cloud infrastructure (where NNNN is the number of your bucket which you can see in the mounted folder /mnt by typing the command df).
includeConfig "$baseDir/params.config"
process.container = 'biocorecrg/c4lwg-2018:latest'
singularity.cacheDir = "$baseDir/singularity"
profiles {
standard {
process {
containerOptions = { workflow.containerEngine == "docker" ? '-u $(id -u):$(id -g)': null}
executor="local"
memory='0.6G'
cpus='1'
time='6h'
withLabel: 'twocpus' {
memory='0.6G'
cpus='1'
}
}
}
cluster {
process {
containerOptions = { workflow.containerEngine == "docker" ? '-u $(id -u):$(id -g)': null}
executor="SGE"
queue = "smallcpus"
memory='1G'
cpus='1'
time='6h'
withLabel: 'twocpus' {
queue = "bigcpus"
memory='4G'
cpus='2'
}
}
}
cloud {
workDir = 's3://nf-class-bucket-XXX/work'
aws.region = 'eu-central-1'
aws.batch.cliPath = '/home/ec2-user/miniconda/bin/aws'
process {
executor = 'awsbatch'
queue = 'spot'
memory='1G'
cpus='1'
time='6h'
withLabel: 'twocpus' {
memory='2G'
cpus='2'
}
}
}
}
We indicate the AWS specific parameters (region and cliPath) and the executor awsbatch. Then we indicate the working directory, that should be mounted as S3 volume. This is mandatory when running Nextflow on the cloud.
We can now launch the pipeline indicating -profile cloud:
nextflow run test3.nf -bg -with-docker -profile cloud > log
Note that there is no longer a work folder in the directory where test3.nf is located, because, in the AWS cloud, the output is copied locally in the folder /mnt/nf-class-bucket-NNN/work (you can see the mounted folder - and the correspondign number - typing df).
The multiqc report can be seen on the AWS webpage at https://nf-class-bucket-NNN.s3.eu-central-1.amazonaws.com/results/ouptut_multiQC/multiqc_report.html
But you need before to change permissions for that file as (where NNNN is the number of your bucket):
chmod 775 /mnt/nf-class-bucket-NNNN/results/ouptut_multiQCmultiqc_report.html
Sometimes you can find that the Nextflow process itself is very memory intensive and the main node can run out of memory. To avoid this, you can reduce the memory needed by setting an environmental variable:
export NXF_OPTS="-Xms50m -Xmx500m"
Again we can copy the output file to the bucket.
We can also tell Nextflow to directly copy the output file to the S3 bucket: to do so, change the parameter outdir in the params file (use the bucket corresponding to your AWS instance):
outdir = "s3://nf-class-bucket-NNNN/results"
EXERCISE
Modify the test3.nf file to make two sub-workflows:
for fastqc of fastq files and bowtie alignment;
for a fastqc analysis of the aligned files produced by bowtie.
For convenience you can use the multiqc config file called config.yaml in the multiqc process.
Solution
#!/usr/bin/env nextflow
/*
* This code enables the new dsl of Nextflow.
*/
nextflow.enable.dsl=2
/*
* NextFlow test pipe
* @authors
* Luca Cozzuto <lucacozzuto@gmail.com>
*
*/
/*
* Input parameters: read pairs
* Params are stored in the params.config file
*/
version = "1.0"
// this prevents a warning of undefined parameter
params.help = false
// this prints the input parameters
log.info """
BIOCORE@CRG - N F TESTPIPE ~ version ${version}
=============================================
reads : ${params.reads}
reference : ${params.reference}
"""
// this prints the help in case you use --help parameter in the command line and it stops the pipeline
if (params.help) {
log.info 'This is the Biocore\'s NF test pipeline'
log.info 'Enjoy!'
log.info '\n'
exit 1
}
/*
* Defining the output folders.
*/
fastqcOutputFolder = "ouptut_fastqc"
alnOutputFolder = "ouptut_aln"
multiqcOutputFolder = "ouptut_multiQC"
/* Reading the file list and creating a "Channel": a queue that connects different channels.
* The queue is consumed by channels, so you cannot re-use a channel for different processes.
* If you need the same data for different processes you need to make more channels.
*/
Channel
.fromPath( params.reads )
.ifEmpty { error "Cannot find any reads matching: ${params.reads}" }
.set {reads}
reference = file(params.reference)
multiconf = file("config.yaml")
/*
* Process 1. Run FastQC on raw data. A process is the element for executing scripts / programs etc.
*/
process fastQC {
publishDir fastqcOutputFolder
tag { "${reads}" }
input:
path reads
output:
path "*_fastqc.*"
script:
"""
fastqc ${reads}
"""
}
/*
* Process 2. Bowtie index
*/
process bowtieIdx {
tag { "${ref}" }
input:
path ref
output:
tuple val("${ref}"), path ("${ref}*.ebwt")
script:
"""
gunzip -c ${ref} > reference.fa
bowtie-build reference.fa ${ref}
rm reference.fa
"""
}
/*
* Process 3. Bowtie alignment
*/
process bowtieAln {
publishDir alnOutputFolder, pattern: '*.sam'
tag { "${reads}" }
label 'twocpus'
input:
tuple val(refname), path (ref_files)
path reads
output:
path "${reads}.sam", emit: samples_sam
path "${reads}.log", emit: samples_log
script:
"""
bowtie -p ${task.cpus} ${refname} -q ${reads} -S > ${reads}.sam 2> ${reads}.log
"""
}
/*
* Process 4. Run multiQC on fastQC results
*/
process multiQC {
publishDir multiqcOutputFolder, mode: 'copy' // this time do not link but copy the output file
input:
path (multiconf)
path (inputfiles)
output:
path("multiqc_report.html")
script:
"""
multiqc . -c ${multiconf}
"""
}
workflow flow1 {
take: reads
main:
fastqc_out = fastQC(reads)
bowtie_index = bowtieIdx(reference)
bowtieAln(bowtie_index, reads)
emit:
sam = bowtieAln.out.samples_sam
logs = bowtieAln.out.samples_log
fastqc_out
}
workflow flow2 {
take: alns
main:
fastqc_out2 = fastQC(alns)
emit:
fastqc_out2
}
workflow {
flow1(reads)
flow2(flow1.out.sam)
multiQC(multiconf, flow1.out.fastqc_out.mix(flow1.out.logs).mix(flow2.out.fastqc_out2).collect())
}
workflow.onComplete {
println ( workflow.success ? "\nDone! Open the following report in your browser --> ${multiqcOutputFolder}/multiqc_report.html\n" : "Oops .. something went wrong" )
}
Modules and how to re-use the code
A great advantage of the new DSL2 is to allow the modularization of the code. In particular, you can move a named workflow within a module and keep it aside for being accessed by different pipelines.
The test4 folder provides an example of using modules.
#!/usr/bin/env nextflow
/*
* This code enables the new dsl of Nextflow.
*/
nextflow.enable.dsl=2
/*
* NextFlow test pipe
* @authors
* Luca Cozzuto <lucacozzuto@gmail.com>
*
*/
/*
* Input parameters: read pairs
* Params are stored in the params.config file
*/
version = "1.0"
// this prevents a warning of undefined parameter
params.help = false
// this prints the input parameters
log.info """
BIOCORE@CRG - N F TESTPIPE ~ version ${version}
=============================================
reads : ${params.reads}
"""
// this prints the help in case you use --help parameter in the command line and it stops the pipeline
if (params.help) {
log.info 'This is the Biocore\'s NF test pipeline'
log.info 'Enjoy!'
log.info '\n'
exit 1
}
/*
* Defining the output folders.
*/
fastqcOutputFolder = "ouptut_fastqc"
multiqcOutputFolder = "ouptut_multiQC"
Channel
.fromPath( params.reads ) // read the files indicated by the wildcard
.ifEmpty { error "Cannot find any reads matching: ${params.reads}" } // if empty, complains
.set {reads_for_fastqc} // make the channel "reads_for_fastqc"
include { multiqc } from "${baseDir}/modules/multiqc" addParams(OUTPUT: multiqcOutputFolder)
include { fastqc } from "${baseDir}/modules/fastqc" addParams(OUTPUT: fastqcOutputFolder, LABEL:"twocpus")
workflow {
fastqc_out = fastqc(reads_for_fastqc)
multiqc(fastqc_out.collect())
}
workflow.onComplete {
println ( workflow.success ? "\nDone! Open the following report in your browser --> ${multiqcOutputFolder}/multiqc_report.html\n" : "Oops .. something went wrong" )
}
We now include two modules, named fastqc and multiqc, from `${baseDir}/modules/fastqc.nf` and `${baseDir}/modules/multiqc.nf`.
Let’s inspect the multiQC module:
/*
* multiqc module
*/
params.CONTAINER = "quay.io/biocontainers/multiqc:1.9--pyh9f0ad1d_0"
params.OUTPUT = "multiqc_output"
process multiqc {
publishDir(params.OUTPUT, mode: 'copy')
container params.CONTAINER
input:
path (inputfiles)
output:
path "multiqc_report.html"
script:
"""
multiqc .
"""
}
The module multiqc takes as input a channel with files containing reads and produces as output the files generated by the multiqc program.
The module contains the directive publishDir, the tag, the container to be used and has similar input, output and script session as the fastqc process in test3.nf.
A module can contain its own parameters that can be used for connecting the main script to some variables inside the module.
In this example we have the declaration of two parameters that are defined at the beginning:
/*
* fastqc module
*/
params.CONTAINER = "quay.io/biocontainers/fastqc:0.11.9--0"
params.OUTPUT = "fastqc_output"
params.LABEL = ""
process fastqc {
publishDir(params.OUTPUT, mode: 'copy')
tag { "${reads}" }
container params.CONTAINER
label (params.LABEL)
input:
path(reads)
output:
path("*_fastqc*")
script:
"""
fastqc -t ${task.cpus} ${reads}
"""
}
They can be overridden from the main script that is calling the module:
The parameter params.OUTPUT can be used for connecting the output of this module with one in the main script.
The parameter params.CONTAINER can be used for declaring the image to use for this particular module.
In this example, in our main script we pass only the OUTPUT parameters by writing them as follows:
#!/usr/bin/env nextflow
/*
* This code enables the new dsl of Nextflow.
*/
nextflow.enable.dsl=2
/*
* NextFlow test pipe
* @authors
* Luca Cozzuto <lucacozzuto@gmail.com>
*
*/
/*
* Input parameters: read pairs
* Params are stored in the params.config file
*/
version = "1.0"
// this prevents a warning of undefined parameter
params.help = false
// this prints the input parameters
log.info """
BIOCORE@CRG - N F TESTPIPE ~ version ${version}
=============================================
reads : ${params.reads}
"""
// this prints the help in case you use --help parameter in the command line and it stops the pipeline
if (params.help) {
log.info 'This is the Biocore\'s NF test pipeline'
log.info 'Enjoy!'
log.info '\n'
exit 1
}
/*
* Defining the output folders.
*/
fastqcOutputFolder = "ouptut_fastqc"
multiqcOutputFolder = "ouptut_multiQC"
Channel
.fromPath( params.reads ) // read the files indicated by the wildcard
.ifEmpty { error "Cannot find any reads matching: ${params.reads}" } // if empty, complains
.set {reads_for_fastqc} // make the channel "reads_for_fastqc"
include { multiqc } from "${baseDir}/modules/multiqc" addParams(OUTPUT: multiqcOutputFolder)
include { fastqc } from "${baseDir}/modules/fastqc" addParams(OUTPUT: fastqcOutputFolder, LABEL:"twocpus")
workflow {
fastqc_out = fastqc(reads_for_fastqc)
multiqc(fastqc_out.collect())
}
workflow.onComplete {
println ( workflow.success ? "\nDone! Open the following report in your browser --> ${multiqcOutputFolder}/multiqc_report.html\n" : "Oops .. something went wrong" )
}
While we keep the information of the container inside the module for better reproducibility:
/*
* multiqc module
*/
params.CONTAINER = "quay.io/biocontainers/multiqc:1.9--pyh9f0ad1d_0"
params.OUTPUT = "multiqc_output"
process multiqc {
publishDir(params.OUTPUT, mode: 'copy')
container params.CONTAINER
input:
path (inputfiles)
output:
path "multiqc_report.html"
script:
"""
multiqc .
"""
}
Here you see that we are not using our own image, but rather we use the image provided by biocontainers in quay.
Let’s have a look at the fastqc.nf module:
/*
* fastqc module
*/
params.CONTAINER = "quay.io/biocontainers/fastqc:0.11.9--0"
params.OUTPUT = "fastqc_output"
params.LABEL = ""
process fastqc {
publishDir(params.OUTPUT, mode: 'copy')
tag { "${reads}" }
container params.CONTAINER
label (params.LABEL)
input:
path(reads)
output:
path("*_fastqc*")
script:
"""
fastqc -t ${task.cpus} ${reads}
"""
}
It is very similar to the multiqc one: we just add an extra parameter for connecting the resources defined in the nextflow.config file and the label indicated in the process. Also in the script part there is a connection between the fastqc command line and the nnumber of threads defined in the nextflow config file.
To use this module, we have to change the main code as follows:
#!/usr/bin/env nextflow
/*
* This code enables the new dsl of Nextflow.
*/
nextflow.enable.dsl=2
/*
* NextFlow test pipe
* @authors
* Luca Cozzuto <lucacozzuto@gmail.com>
*
*/
/*
* Input parameters: read pairs
* Params are stored in the params.config file
*/
version = "1.0"
// this prevents a warning of undefined parameter
params.help = false
// this prints the input parameters
log.info """
BIOCORE@CRG - N F TESTPIPE ~ version ${version}
=============================================
reads : ${params.reads}
"""
// this prints the help in case you use --help parameter in the command line and it stops the pipeline
if (params.help) {
log.info 'This is the Biocore\'s NF test pipeline'
log.info 'Enjoy!'
log.info '\n'
exit 1
}
/*
* Defining the output folders.
*/
fastqcOutputFolder = "ouptut_fastqc"
multiqcOutputFolder = "ouptut_multiQC"
Channel
.fromPath( params.reads ) // read the files indicated by the wildcard
.ifEmpty { error "Cannot find any reads matching: ${params.reads}" } // if empty, complains
.set {reads_for_fastqc} // make the channel "reads_for_fastqc"
include { multiqc } from "${baseDir}/modules/multiqc" addParams(OUTPUT: multiqcOutputFolder)
include { fastqc } from "${baseDir}/modules/fastqc" addParams(OUTPUT: fastqcOutputFolder, LABEL:"twocpus")
workflow {
fastqc_out = fastqc(reads_for_fastqc)
multiqc(fastqc_out.collect())
}
workflow.onComplete {
println ( workflow.success ? "\nDone! Open the following report in your browser --> ${multiqcOutputFolder}/multiqc_report.html\n" : "Oops .. something went wrong" )
}
The label twocpus is specified in the nextflow.config file for each profile:
Note
IMPORTANT: You have to specify a default image to run nextflow -with-docker or -with-singularity and you have to have a container(s) defined inside modules.
EXERCISE
Make a module wrapper for the bowtie tool and change the script in test3 accordingly.
Solution
Solution in the folder test5
Reporting and graphical interface
Nextflow has an embedded function for reporting informations about the resources requested for each job and the timing; to generate a html report, run Nextflow with the -with-report parameter :
nextflow run test5.nf -with-docker -bg -with-report > log
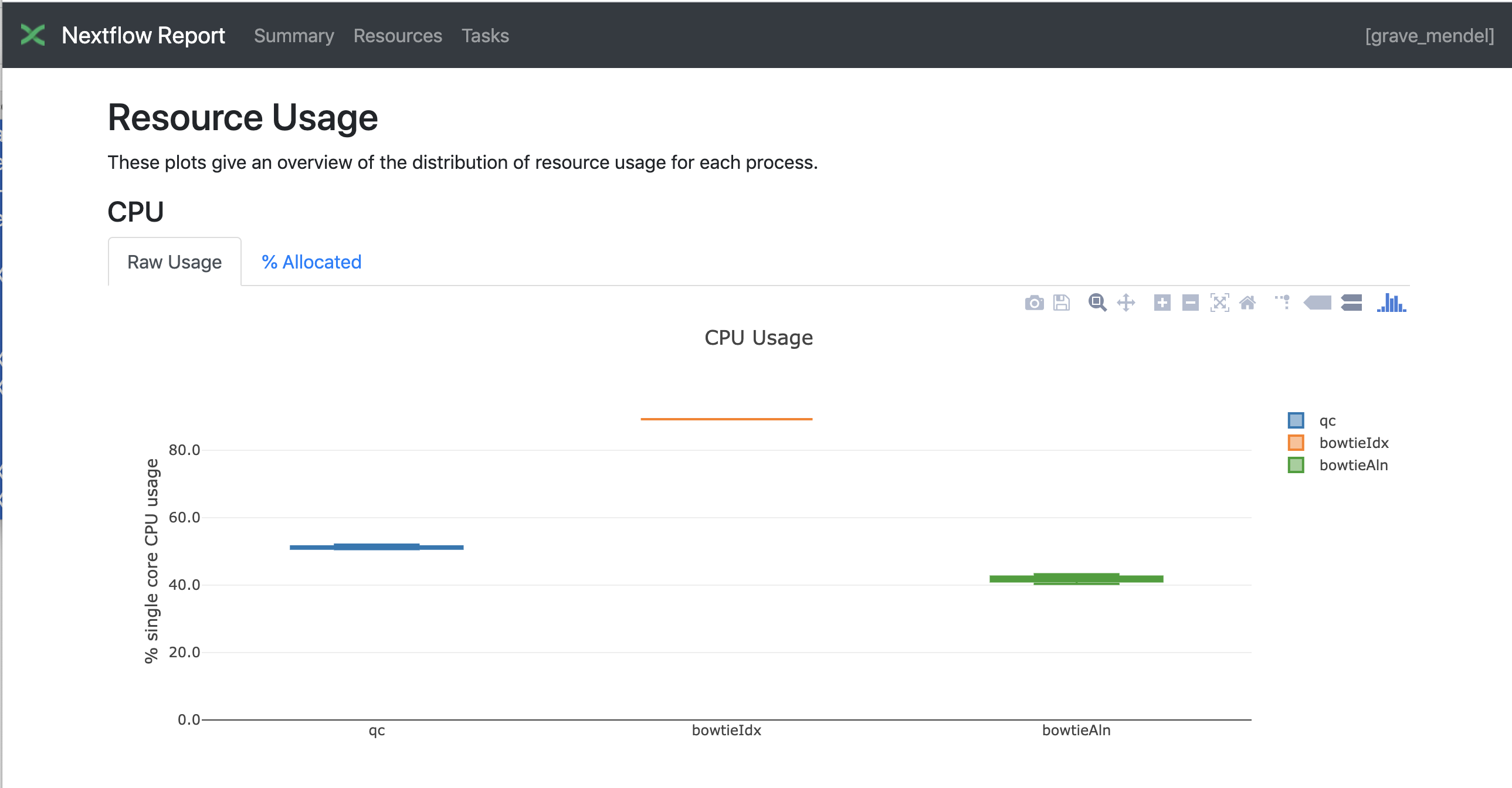
Nextflow Tower is an open source monitoring and managing platform for Nextflow workflows. There are two versions:
Open source for monitoring of single pipelines.
Commercial one for workflow management, monitoring and resource optimisation.
We will show the open source one.
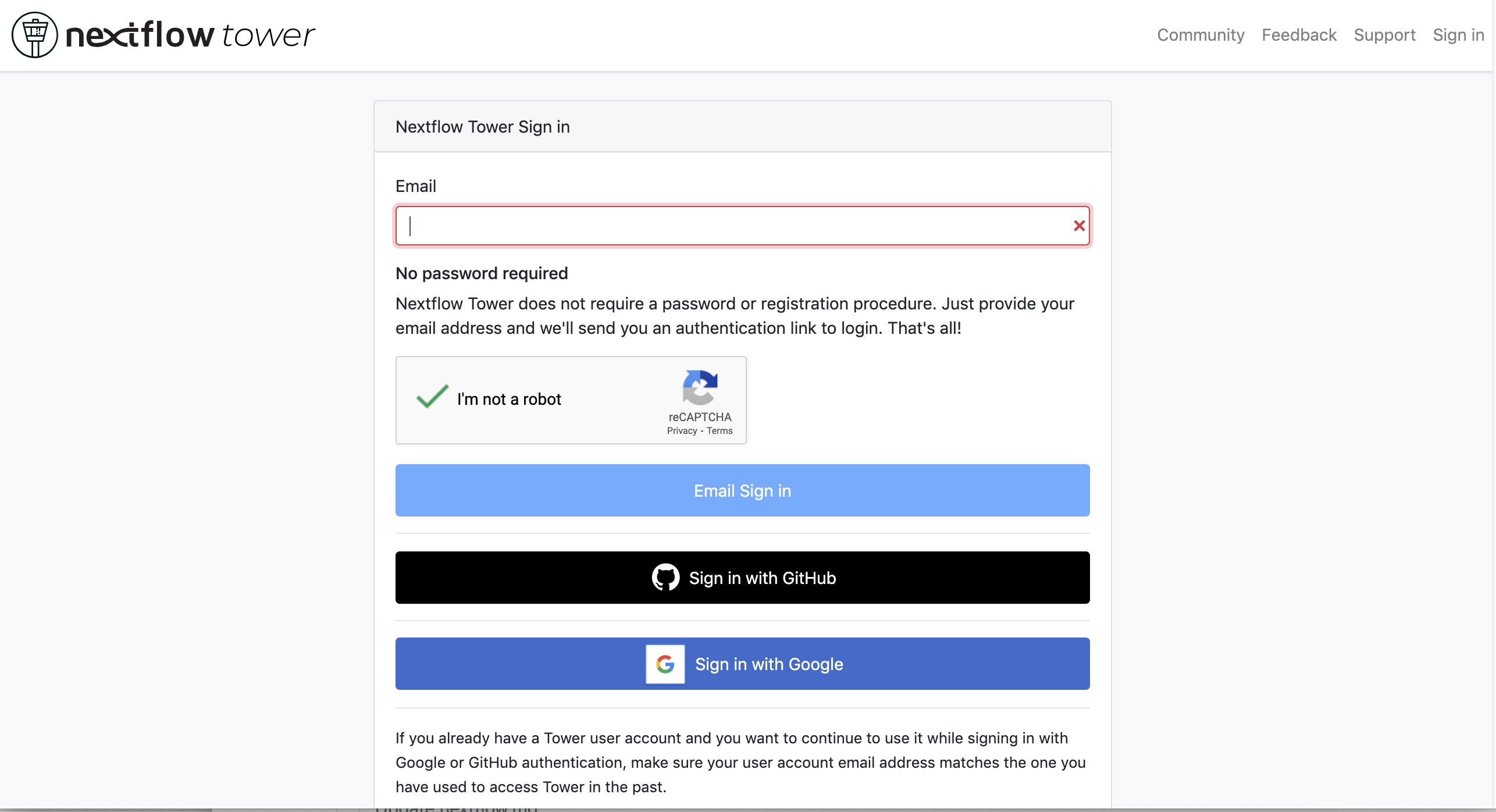
First, you need to access the tower.nf website and login.

If you selected the email for receiving the instructions and the token to be used.
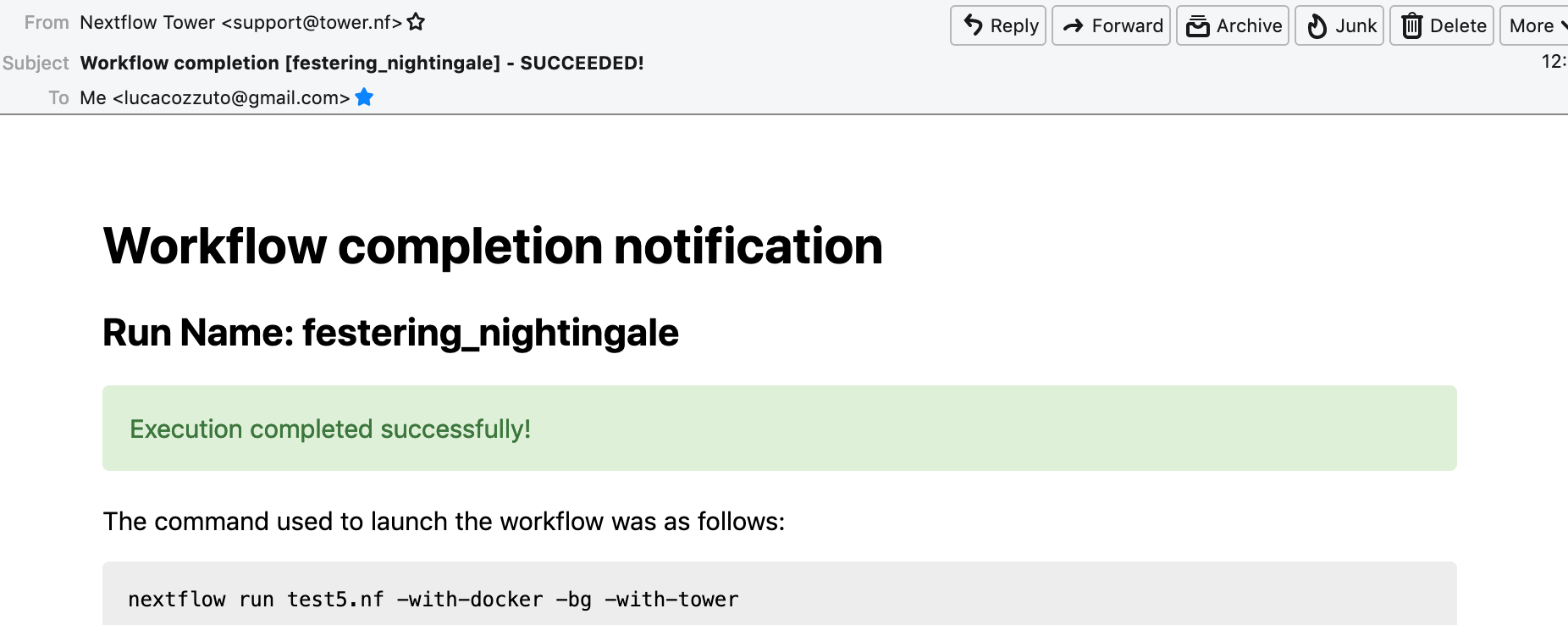
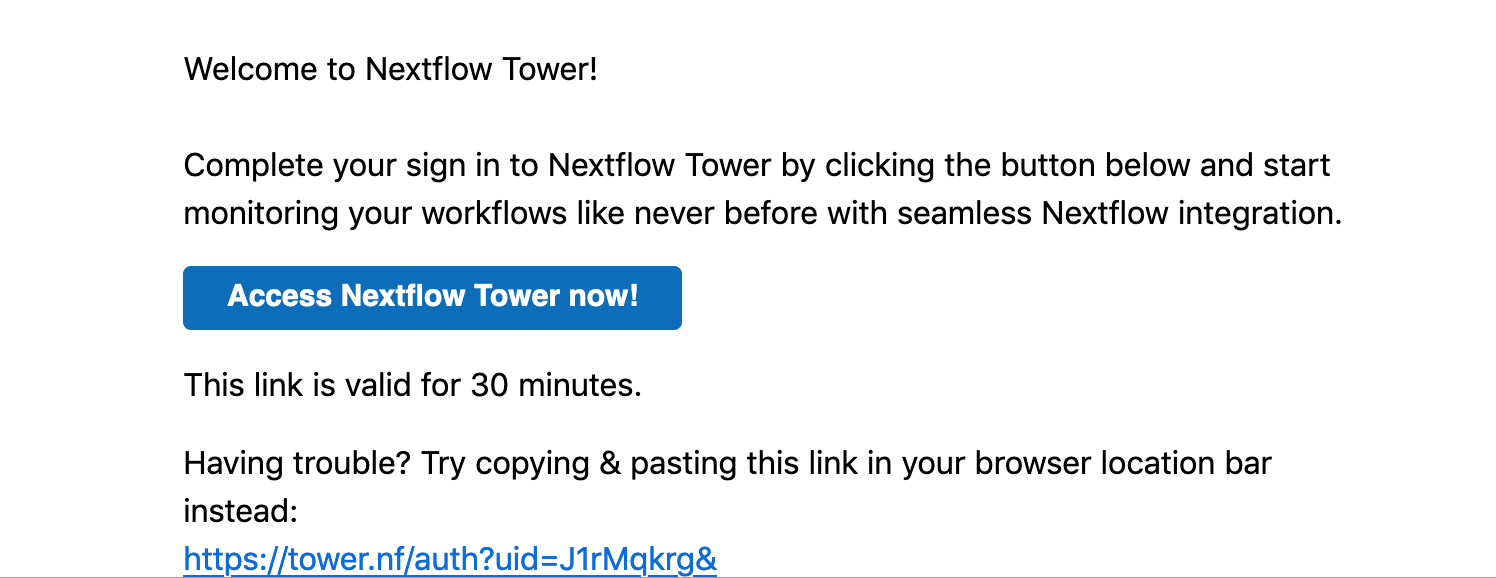
check the email:
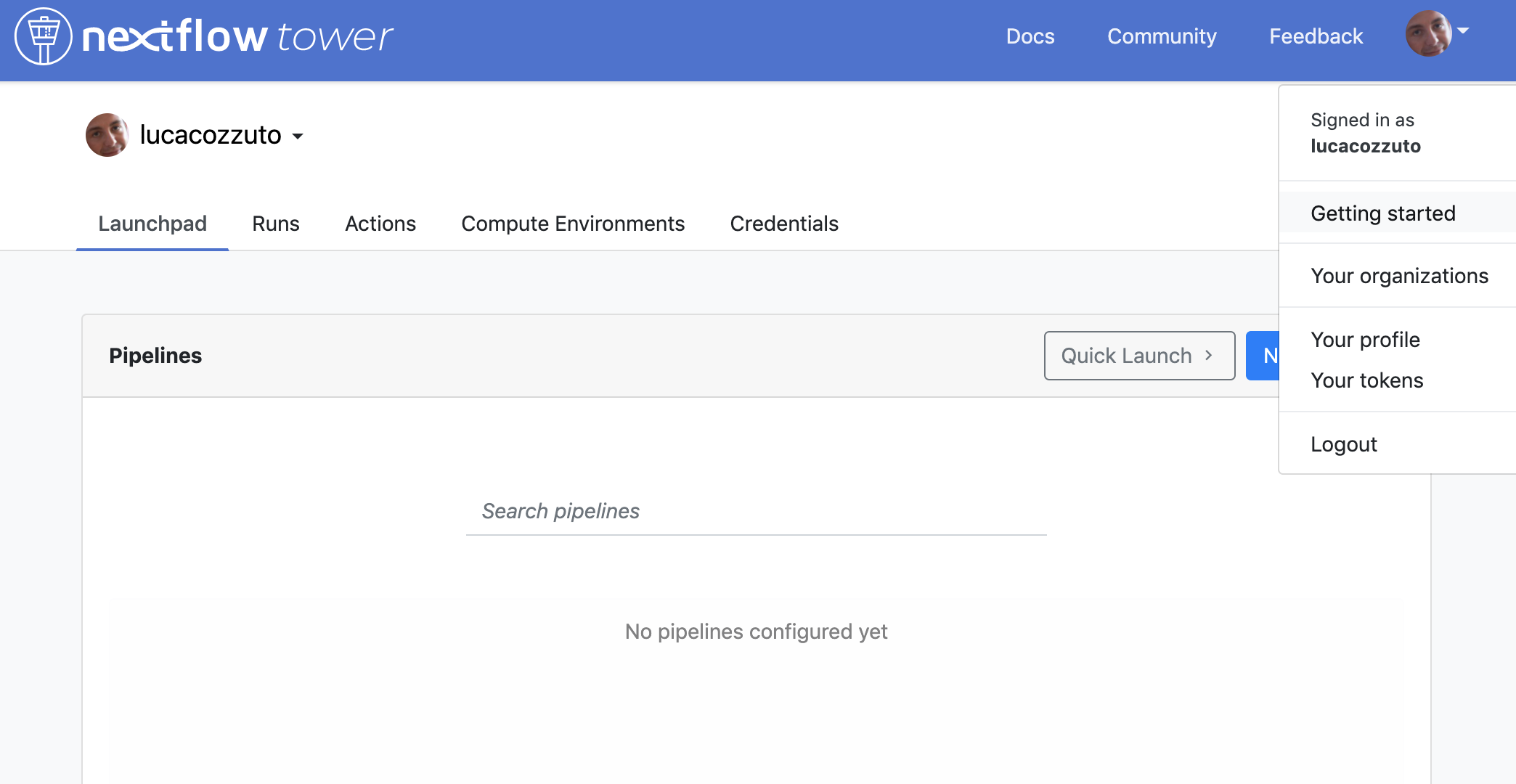
You can generate your token at https://tower.nf/tokens and copy paste it in your pipeline using this snippet in the configuration file:
tower {
accessToken = '<YOUR TOKEN>'
enabled = true
}
or exporting those environmental variables:
export TOWER_ACCESS_TOKEN=*******YOUR***TOKEN*****HERE*******
export NXF_VER=21.04.0
Now we can launch the pipeline:
nextflow run test5.nf -with-singularity -with-tower -bg > log
CAPSULE: Downloading dependency io.nextflow:nf-tower:jar:20.09.1-edge
CAPSULE: Downloading dependency org.codehaus.groovy:groovy-nio:jar:3.0.5
CAPSULE: Downloading dependency io.nextflow:nextflow:jar:20.09.1-edge
CAPSULE: Downloading dependency io.nextflow:nf-httpfs:jar:20.09.1-edge
CAPSULE: Downloading dependency org.codehaus.groovy:groovy-json:jar:3.0.5
CAPSULE: Downloading dependency org.codehaus.groovy:groovy:jar:3.0.5
CAPSULE: Downloading dependency io.nextflow:nf-amazon:jar:20.09.1-edge
CAPSULE: Downloading dependency org.codehaus.groovy:groovy-templates:jar:3.0.5
CAPSULE: Downloading dependency org.codehaus.groovy:groovy-xml:jar:3.0.5
and go to the tower website again:
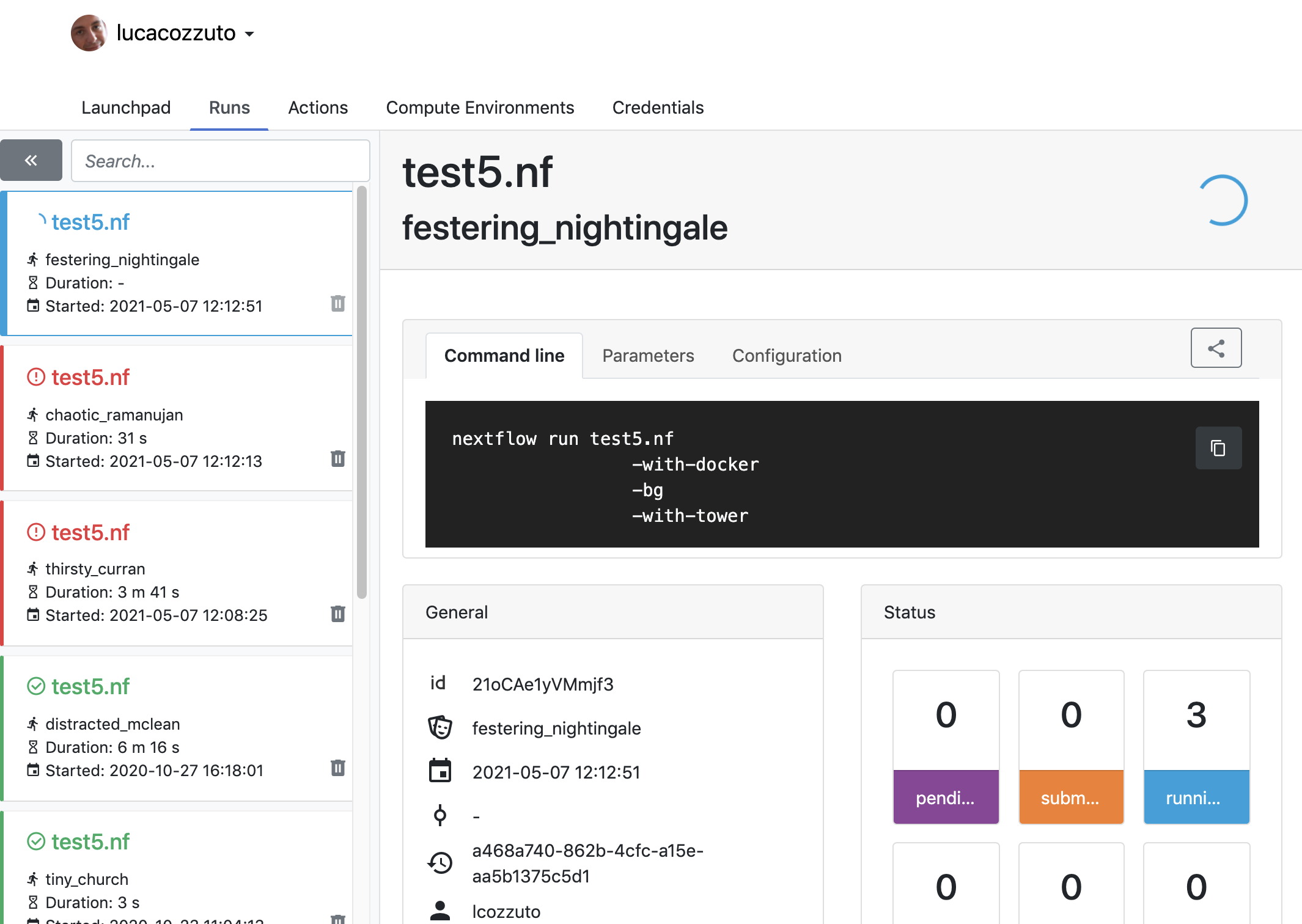
When the pipeline is finished we can also receive a mail.