Singularity
Intoduction to Singularity
- Focus:
- Reproducibility to scientific computing and the high-performance computing (HPC) world.
- Origin: Lawrence Berkeley National Laboratory. Later spin-off: Sylabs
- Version 1.0 -> 2016
- More information: https://en.wikipedia.org/wiki/Singularity_(software)
Singularity architecture
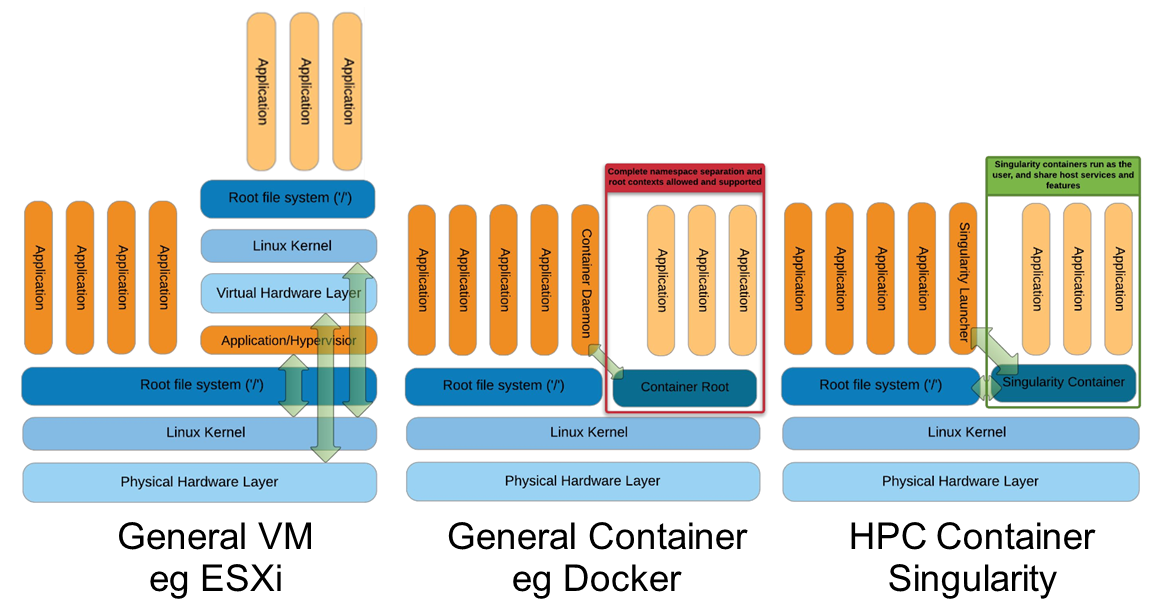
Strengths
- No dependency of a daemon
- Can be run as a simple user
- Avoid permission headaches and hacks
- Image/container is a file (or directory)
- More easily portable
- Two type of images
- Read-only (production)
- Writable (development, via sandbox)
Weaknesses
- At the time of writing only good support in Linux
- Mac experimental. Desktop edition. Only running
- For some features you need root account (or sudo)
Build process
Examples
Registries
- Docker Hub
https://hub.docker.com/r/biocontainers/fastqc
singularity build fastqc-0.11.9_cv7.sif docker://biocontainers/fastqc:v0.11.9_cv7
- Biocontainers
Via quay.io
https://quay.io/repository/biocontainers/fastqc
singularity build fastqc-0.11.9.sif docker://quay.io/biocontainers/fastqc:0.11.9--0
Via Galaxy project prebuilt images
singularity pull --name fastqc-0.11.9.sif https://depot.galaxyproject.org/singularity/fastqc:0.11.9--0
-
Docker Daemon (local Docker image)
singularity build fastqc-web-0.11.9.sif docker-daemon://fastqcwww -
Also Singularity registries (not so popular so far)
Sandboxing
singularity build --sandbox ./sandbox docker://ubuntu:18.04
touch sandbox/etc/myetc.conf
singularity build sandbox.sif ./sandbox
Singularity recipes
Docker bootstrap
BootStrap: docker
From: biocontainers/fastqc:v0.11.9_cv7
%runscript
echo "Welcome to FastQC Image"
fastqc --version
%post
echo "Image built"
sudo singularity build fastqc.sif docker.singularity
Debian bootstrap
BootStrap: debootstrap
OSVersion: bionic
MirrorURL: http://fr.archive.ubuntu.com/ubuntu/
Include: build-essential curl python python-dev openjdk-11-jdk bzip2 zip unzip
%runscript
echo "Welcome to my Singularity Image"
fastqc --version
multiqc --version
bowtie --version
%post
FASTQC_VERSION=0.11.9
MULTIQC_VERSION=1.9
BOWTIE_VERSION=1.3.0
cd /usr/local; curl -k -L https://www.bioinformatics.babraham.ac.uk/projects/fastqc/fastqc_v${FASTQC_VERSION}.zip > fastqc.zip
cd /usr/local; unzip fastqc.zip; rm fastqc.zip; chmod 775 FastQC/fastqc; ln -s /usr/local/FastQC/fastqc /usr/local/bin/fastqc
cd /usr/local; curl --fail --silent --show-error --location --remote-name https://github.com/BenLangmead/bowtie/releases/download/v$BOWTIE_VERSION/bowtie-${BOWTIE_VERSION}-linux-x86_64.zip
cd /usr/local; unzip -d /usr/local bowtie-${BOWTIE_VERSION}-linux-x86_64.zip
cd /usr/local; rm bowtie-${BOWTIE_VERSION}-linux-x86_64.zip
cd /usr/local/bin; ln -s ../bowtie-${BOWTIE_VERSION}-linux-x86_64/bowtie* .
curl --fail --silent --show-error --location --remote-name https://bootstrap.pypa.io/get-pip.py
python get-pip.py
pip install numpy matplotlib
pip install -I multiqc==${MULTIQC_VERSION}
echo "Biocore image built"
%labels
Maintainer Biocorecrg
Version 0.1.0
sudo singularity build fastqc-multi-bowtie.sif debootstrap.singularity
Remote building (optional)
Actually possible with Singularity account. More details at https://sylabs.io/guides/3.6/user-guide/cloud_library.html
singularity remote login
Create account and tokenfile at https://cloud.sylabs.io/
singularity remote login --tokenfile mytokenfile
Build an image
singularity build --remote focal.sif docker://ubuntu:focal
Run and execution process
Singularity shell
Interactive shell.
singularity shell fastqc-multi-bowtie.sif
Singularity run
Execute runscript from recipe definition. Not so common for HPC uses. More for instances (servers).
singularity run fastqc-multi-bowtie.sif
Singularity exec
Execute commands. Common approach in HPC uses.
singularity exec fastqc-multi-bowtie.sif fastqc
Environment control
singularity shell -e fastqc-multi-bowtie.sif
singularity run -e fastqc-multi-bowtie.sif
singularity exec -e fastqc-multi-bowtie.sif fastqc
Compare env command with and without -e modifier.
singularity exec fastqc-multi-bowtie.sif env
singularity exec -e fastqc-multi-bowtie.sif env
Execute from sandboxed images / directories
singularity exec ./sandbox ls -l /etc/myetc.conf
# We can see file created in the directory before
singularity exec ./sandbox bash -c 'apt-get update && apt-get install python'
# We cannot install python
singularity exec --writable ./sandbox bash -c 'apt-get update && apt-get install python'
# We needed to add writable parameter
Run straight from registry
singularity exec docker://ncbi/blast:2.10.1 blastp -version
Bind paths (aka volumes)
Paths of host system mounted in the container
- Default ones, no need to mount them explicitly (for 3.6.x):
$HOME,/sys:/sys,/proc:/proc,/tmp:/tmp,/var/tmp:/var/tmp,/etc/resolv.conf:/etc/resolv.conf,/etc/passwd:/etc/passwd, and$PWDhttps://sylabs.io/guides/3.6/user-guide/bind_paths_and_mounts.html
For others, need to be done explicitly (syntax: host:container)
mkdir testdir
touch testdir/testout
singularity shell -e -B ./testdir:/scratch fastqc-multi-bowtie.sif
> touch /scratch/testin
> exit
ls -l testdir
Exercises:
Using the 2 fastqc available files, process them outside and inside the mounted directory.
Instances
Also know as services. Despite Docker it is still more convenient for these tasks, it allows enabling thing such as webservices (e.g., via APIs) in HPC workflows.
Simple example:
Bootstrap: docker
From: library/mariadb:10.3
%startscript
mysqld
sudo singularity build mariadb.sif mariadb.singularity
mkdir -p testdir
mkdir -p testdir/db
mkdir -p testdir/socket
singularity exec -B ./testdir/db:/var/lib/mysql mariadb.sif mysql_install_db
singularity instance start -B ./testdir/db:/var/lib/mysql -B ./testdir/socket:/run/mysqld mariadb.sif mydb
singularity instance list
singularity exec instance://mydb mysql -uroot
singularity instance stop mydb
More information:
- https://sylabs.io/guides/3.6/user-guide/running_services.html
- https://sylabs.io/guides/3.6/user-guide/networking.html
Troubleshooting
singularity --help
Fakeroot
Singularity permissions are an evolving field. If you don’t have access to sudo, it might be worth considering using –fakeroot/-f parameter.
- More details at https://sylabs.io/guides/3.6/user-guide/fakeroot.html
Singularity cache directory
$HOME/.singularity
- It stores cached images from registries, instances, etc.
- If problems may be a good place to clean. When running
sudo, $HOME is /root.
Global singularity configuration
Normally at /etc/singularity/singularity.conf or similar (e.g preceded by /usr/local/)
- It can only be modified by users with administration permissions
- Worth noting
bind pathlines, which point default mounted directories in containers
| Previous page | Home | Next page |



