Functional analysis
Data bases
Gene Ontology

The Gene Ontology (GO) describes our knowledge of the biological domain with respect to three aspects:
| GO domains / root terms | Description |
|---|---|
| Molecular Function | Molecular-level activities performed by gene products. e.g. catalysis, binding. |
| Biological Process | Larger processes accomplished by multiple molecular activities. e.g. apoptosis, DNA repair. |
| Cellular Component | The locations where a gene product performs a function. e.g. cell membrane, ribosome. |
Example of GO annotation: the gene product “cytochrome c” can be described by:
- the molecular function oxidoreductase activity
- the biological process oxidative phosphorylation
- the cellular component mitochondrial matrix.
The structure of GO can be described as a graph: each GO term is a node, each edge represents the relationships between the nodes. For example:
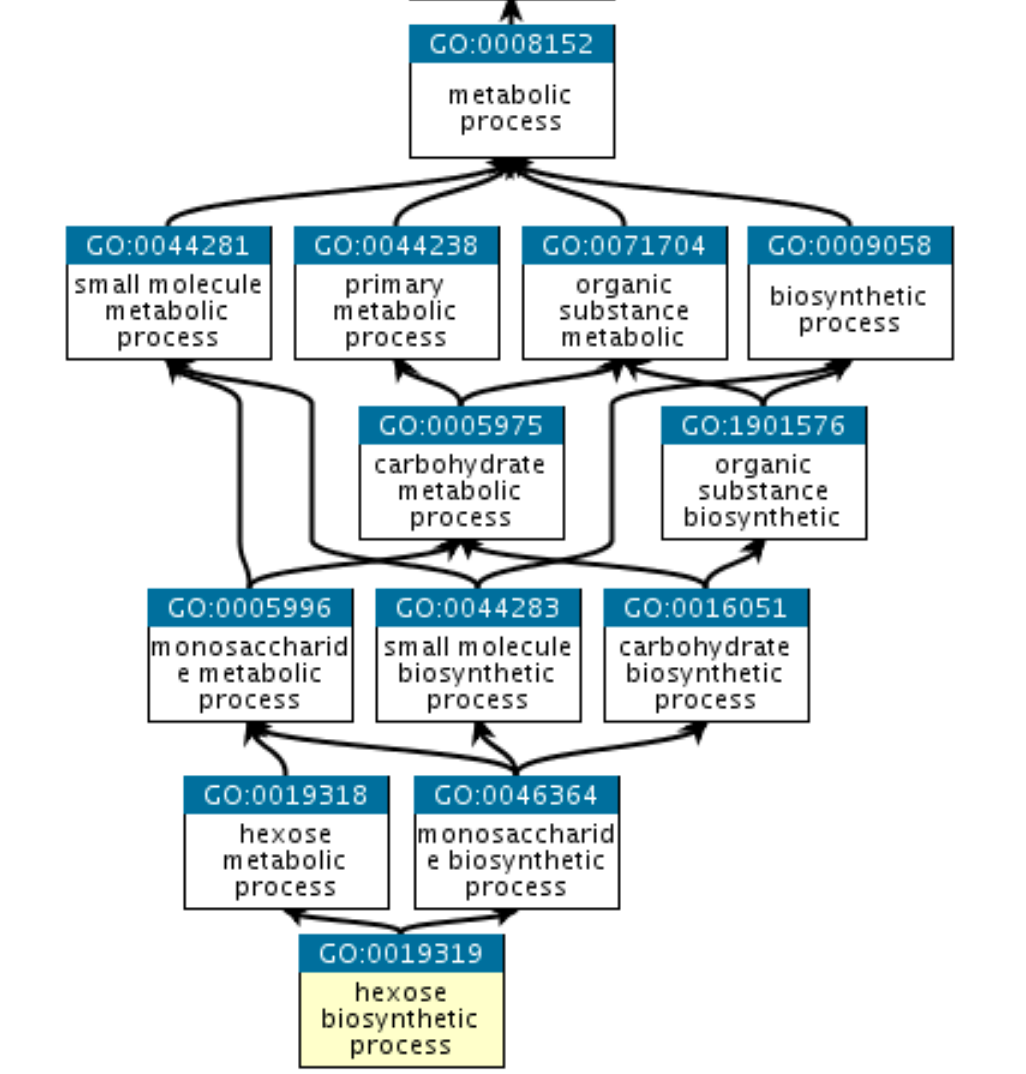
GO:0019319 (hexose biosynthetic process) is part of GO:0019318 (hexose metabolic process) and also part of GO:0046364 (monosaccharide biosynthetic process).
They all share common parent nodes, for example GO:0008152 (metabolic process), and eventually a root node that is here biological process.
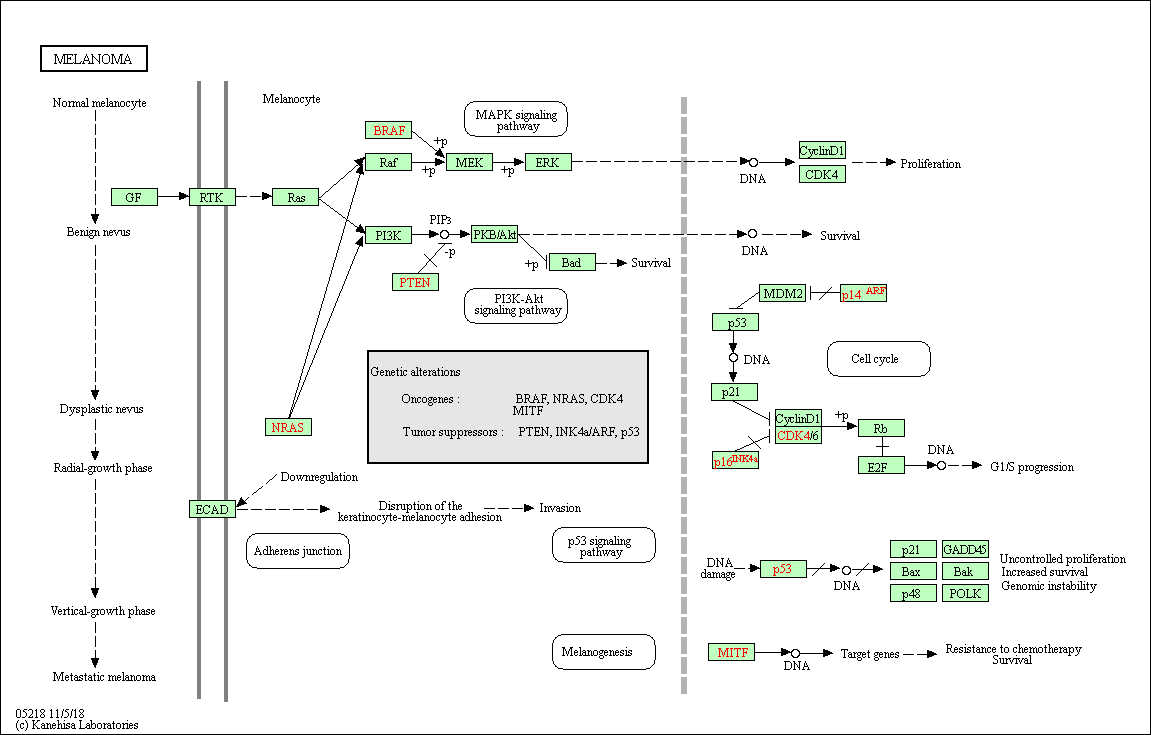
KEGG pathways
The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database for understanding high-level functions and utilities of the biological system.
It provides comprehensible manually-drawn pathways representing biological processes or disease-specific pathways.
Example of the Homo sapiens melanoma pathway:
Molecular Signatures Database (MSigDB)
The Molecular Signatures Database (MSigDB) is a collection of 17810 annotated gene sets (as of May 2019) created to be used with the GSEA software (but not only).
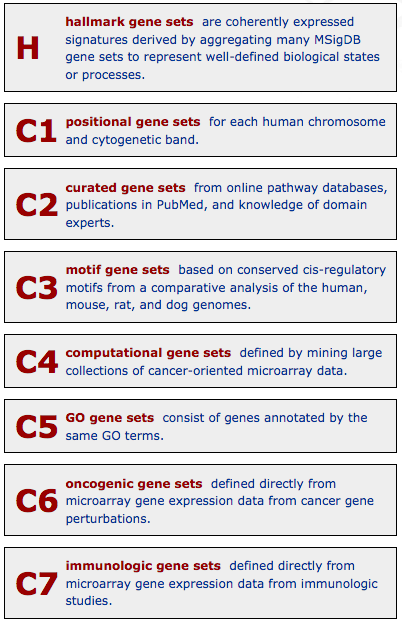
It is divided into 8 major collections (that include the previously described Gene Ontologies and KEGG pathways):
Methods for functional enrichment
Enrichment analysis based on gene selection
Tools based on a user-selection of genes usually require 2 inputs:
- Gene Universe (optional): all genes present in a system: in the case of a high throughput assay, it would be all genes used in the analysis/annotation
- List of genes selected from the universe (required): based on differential gene expression results. For example, adjusted / corrected p-value < 0.05.
They are often based on the Hypergeometric test or on the Fisher’s exact test. You can have a look at this page for some explanation of both tests.
EnrichR
EnrichR is a gene-list enrichment tool developped at the Icahn Schoold of Medicine (Mount Sinai).
It does not require the input of a gene universe: only a selection of genes or a BED file.
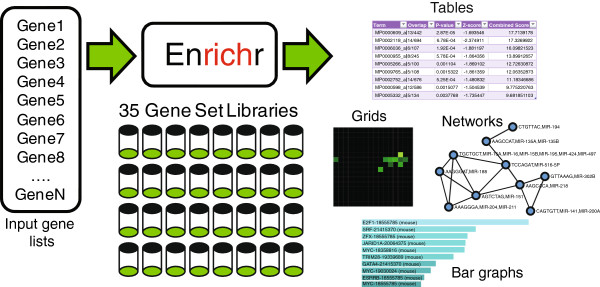
The default EnrichR interface works for Homo sapiens and Mus musculus.
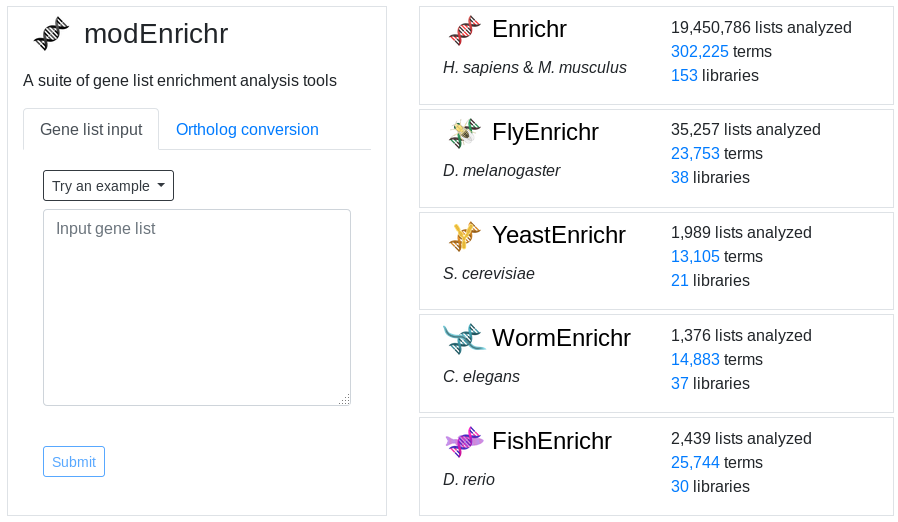
However, EnrichR also provides a set of tools for ortholog conversion and enrichment analysis of more organisms:
In the main page, click on Try a gene set example to get a test list of genes. And Submit.
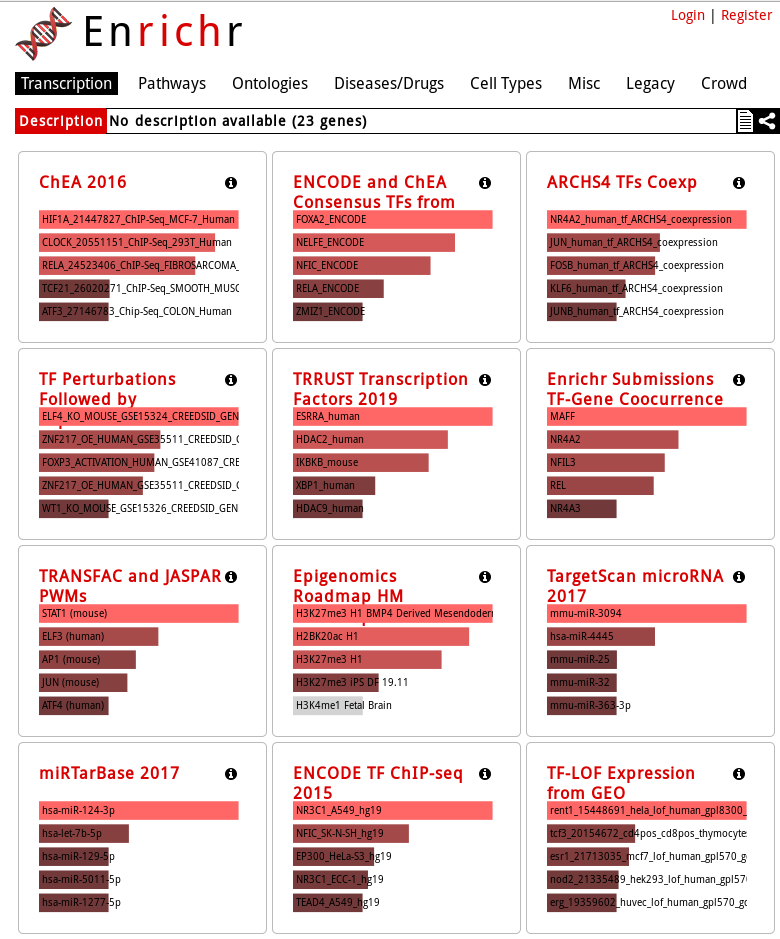
EnrichR tests enrichment of a lot of different gene sets, organized in 8 categories:
- Transcription
- Pathways
- Ontologies
- Diseases/Drugs
- Cell Types
- Misc
- Legacy
- Crowd
More details about each data base analysed can be found here
Now click on Pathways and then enter KEGG 2019 Human.
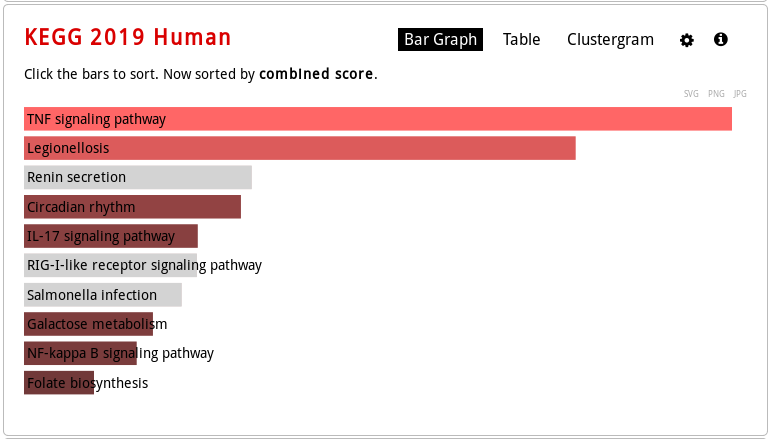
KEGG Human pathway bar graph visualization (mouse over to see the enrichment statistics):
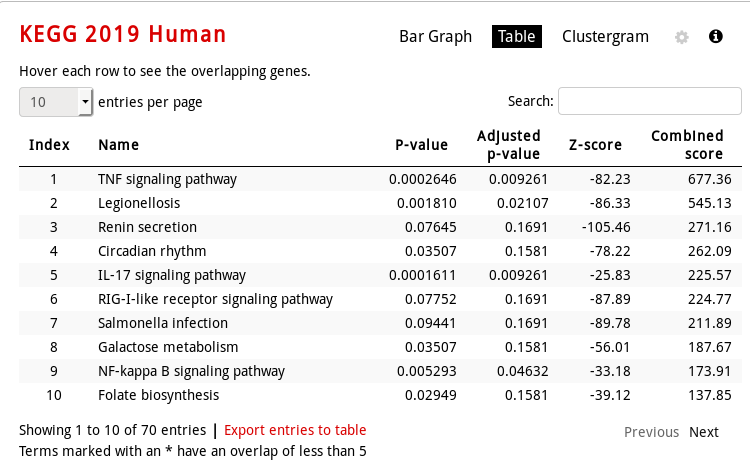
KEGG Human pathway table visualization (mouse over a row to see the genes involved):
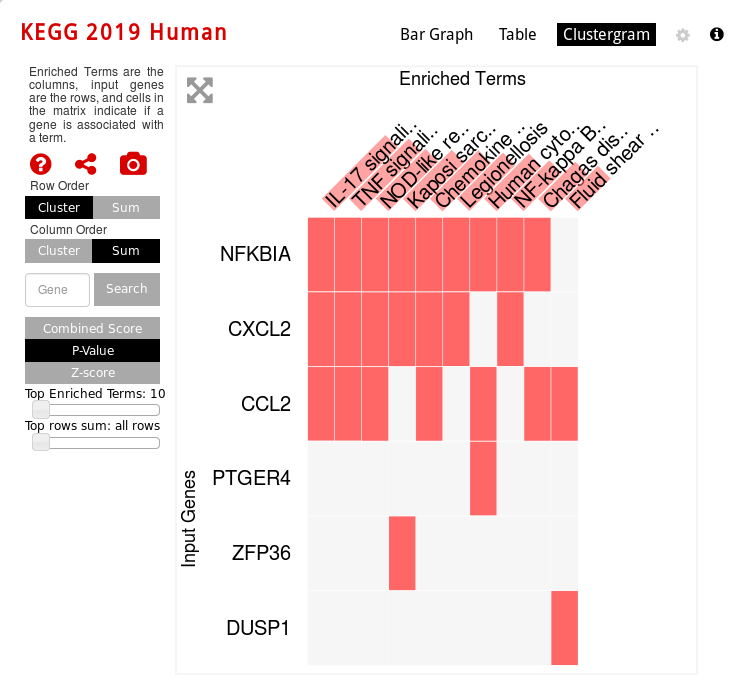
KEGG Human pathway clustergram vizualization (which genes are involved in which enriched pathway):
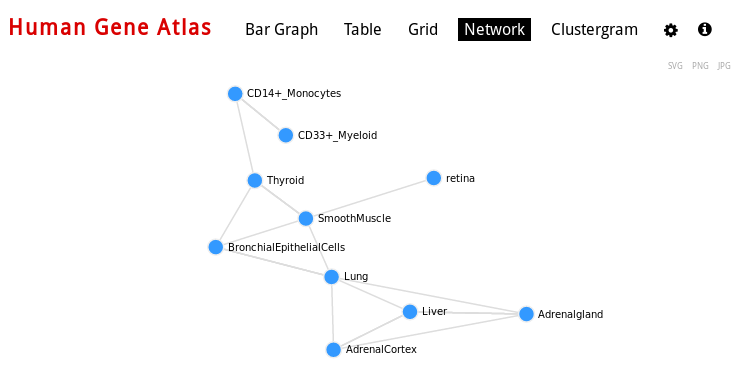
For Cell Types, you can also visualize networks, for example Human gene Atlas:
You can also export some graphs as PNG, JPEG or SVG.
Enrichment based on ranking: GSEA (Gene Set Enrichment Analysis)
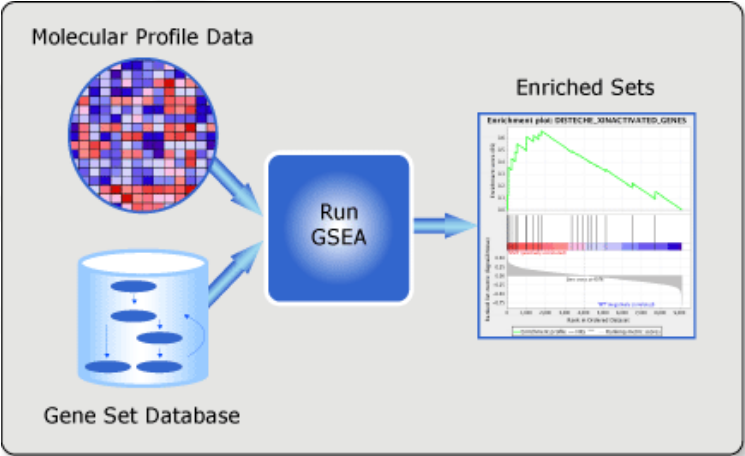
GSEA is available as a Java-based tool.
Algorithm
GSEA doesn’t require a threshold: the whole set of genes is considered.
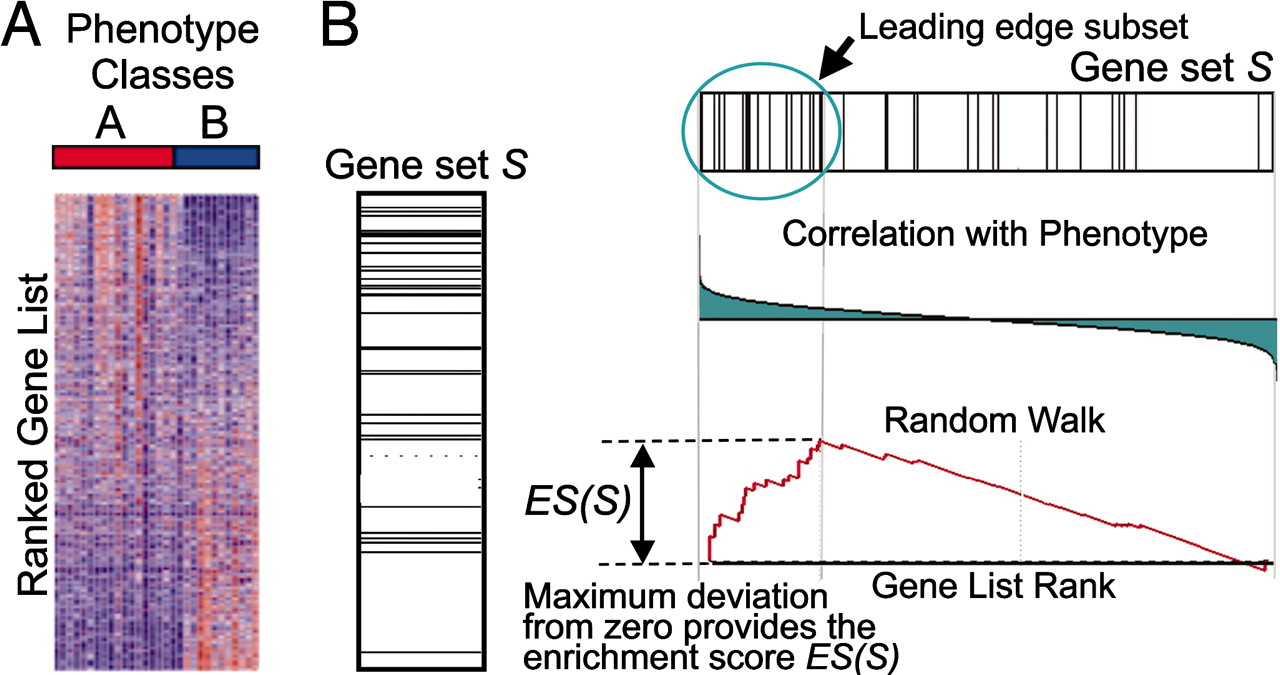
GSEA checks whether a particular gene set (for example, a gene ontology) is randomly distributed across a list of ranked genes.
The algorithm consists of 3 key elements:
- Calculation of the Enrichment Score The Enrichment Score (ES) reflects the degree to which a gene set is overrepresented at the extremes (top or bottom) of the entire ranked gene list.
- Estimation of Significance Level of ES The statistical significant (nominal p-value) of the Enrichment Score (ES) is estimated by using an empirical phenotype-based permutation test procedure. The Normalized Enrichment Score (NES) is obtained by normalizing the ES for each gene set to account for the size of the set.
- Adjustment for Multiple Hypothesis Testing Calculation of the FDR ti control the proportion of falses positives.
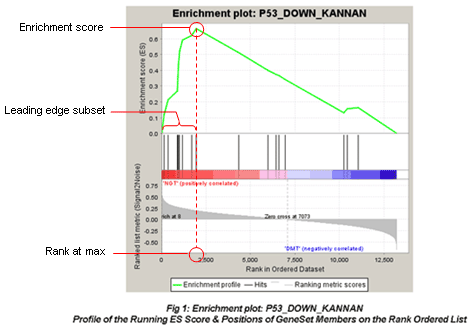
See the GSEA Paper for more details on the algorithm.
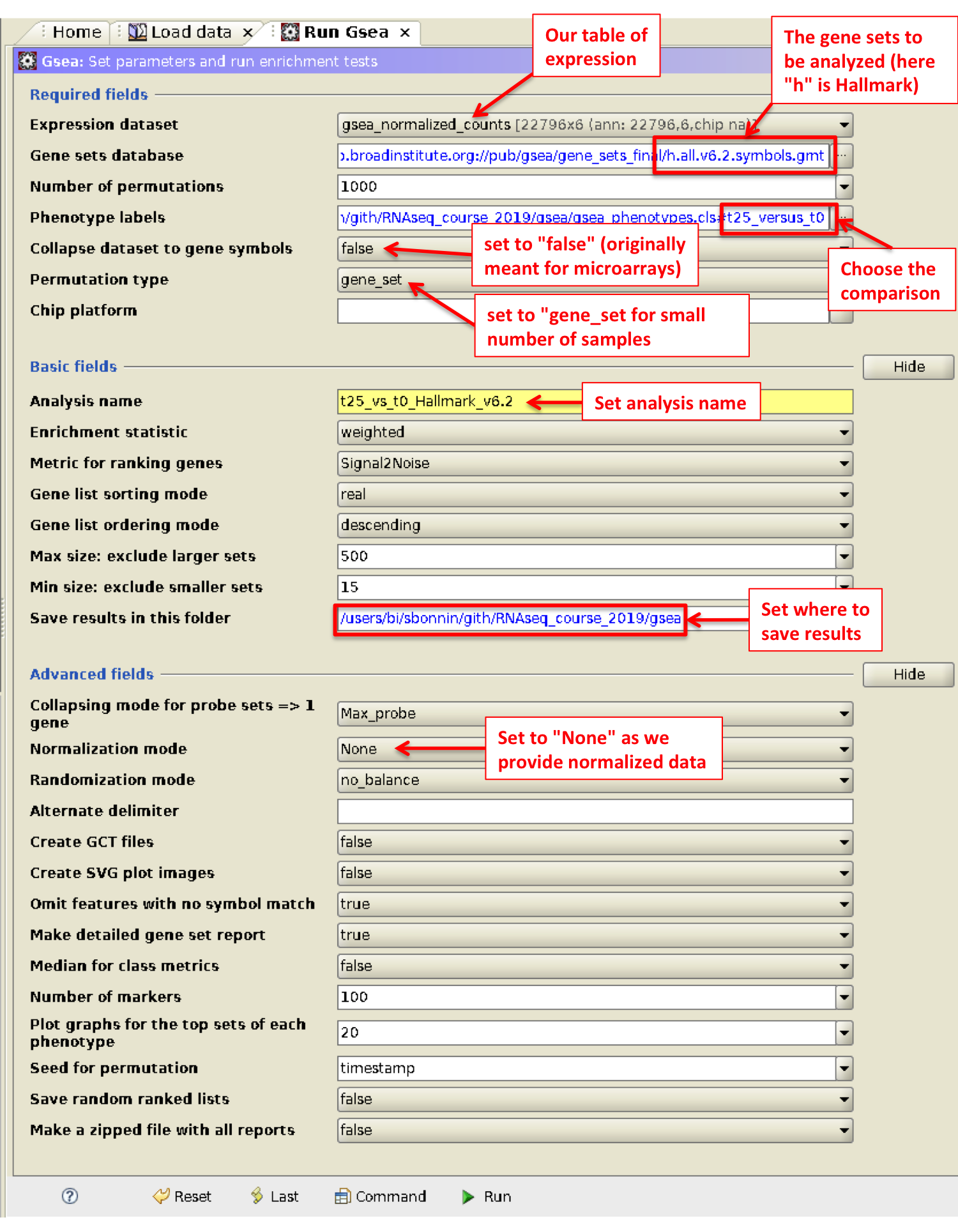
The main GSEA algorithm requires 3 inputs:
- Gene expression data
- Phenotype labels
- Gene sets
Gene expression data in TXT format
In the case an RNA-sequencing experiment, the input should be normalized read counts.
- The first column contains the gene symbols (HUGO symbols for Homo sapiens).
- The second column contains any description or other IDs/symbols, and will be ignored by the algorithm (it will be given in the output).
- The remaining columns contains normalized expressions: one column per sample.
| NAME | DESCRIPTION | WT1 | WT2 | KO1 | KO2 | KO3 |
| DKK1 | NA | 0 | 0 | 0 | 0 | 0 |
| HGT | NA | 0 | 0 | 0 | 0 | 0 |
Phenotype labels in CLS format
A phenotype label file defines phenotype labels (experimental groups) and assigns those labels to the samples in the corresponding expression data file.
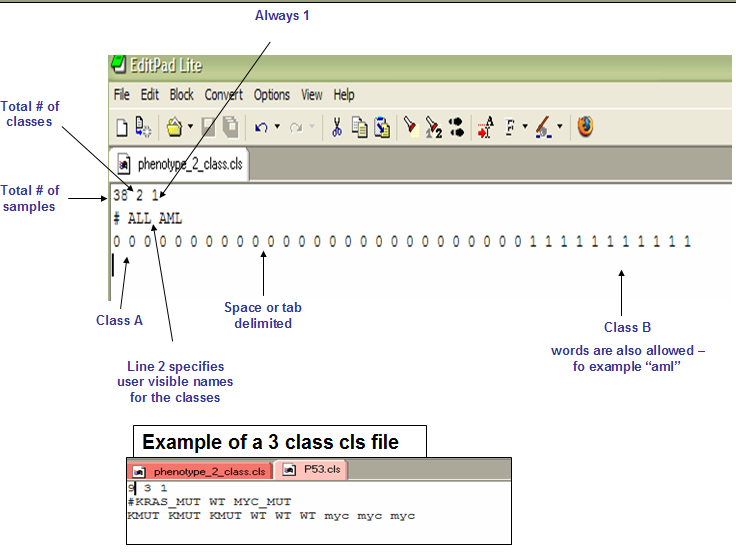
For our example, the file would be like:
| 5 | 2 | 1 | ||
| # | WT | KO | ||
| WT | WT | KO | KO | KO |
NOTE: the first label used is assigned to the first class named on the second line; the second unique label is assigned to the second class named; and so on.
So the phenotype file could also be:
| 5 | 2 | 1 | ||
| # | WT | KO | ||
| 0 | 0 | 1 | 1 | 1 |
The first label WT in the second line is associated to the first label 0 on the third line.
Get the data we will try to run GSEA with:
Download and extract this archive
Download and run GSEA
Download Java application:
Enter the registration page, enter your email and organization, then to download page, enter your Email and login:
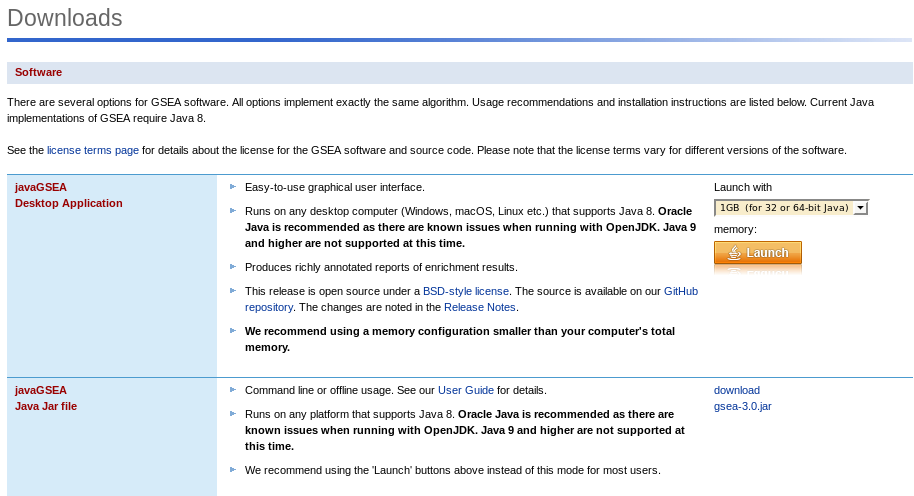
Click on Launch in the GSEA v4.0.3 Java Web Start
(all platforms) section (You need a JAVA 8 installation).
Configure
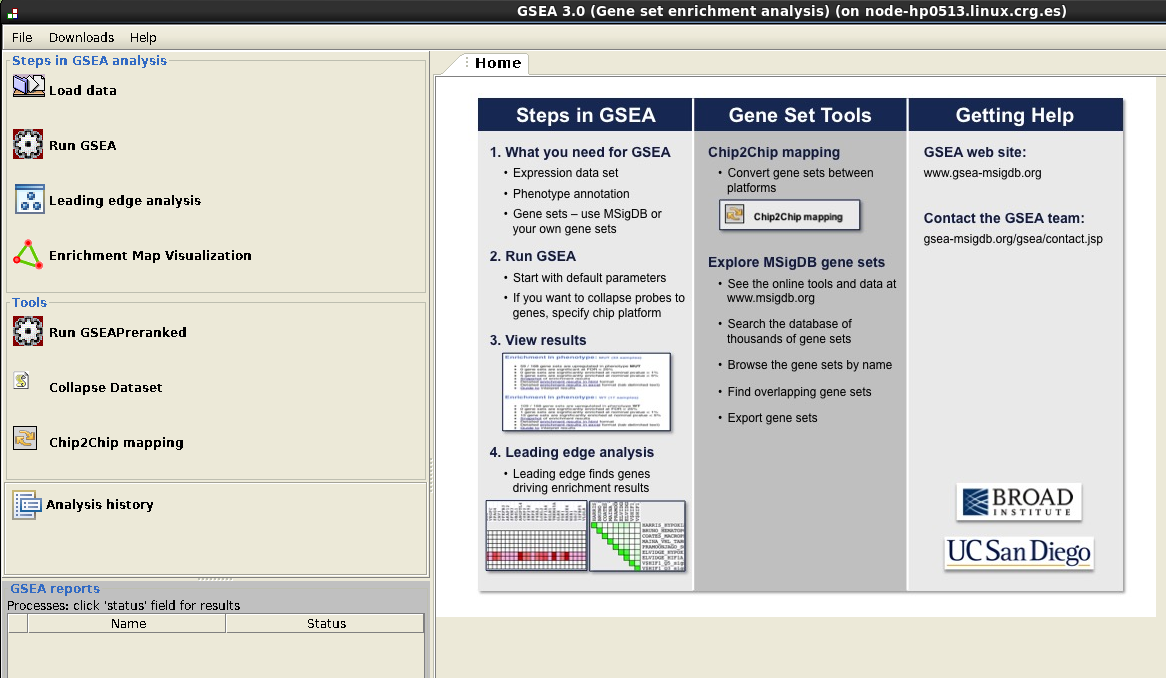
In Steps in GSEA analysis (upper left corner):
- Go to Load data: select normalized_counts_log2.txt and phenotypes.cls and load.
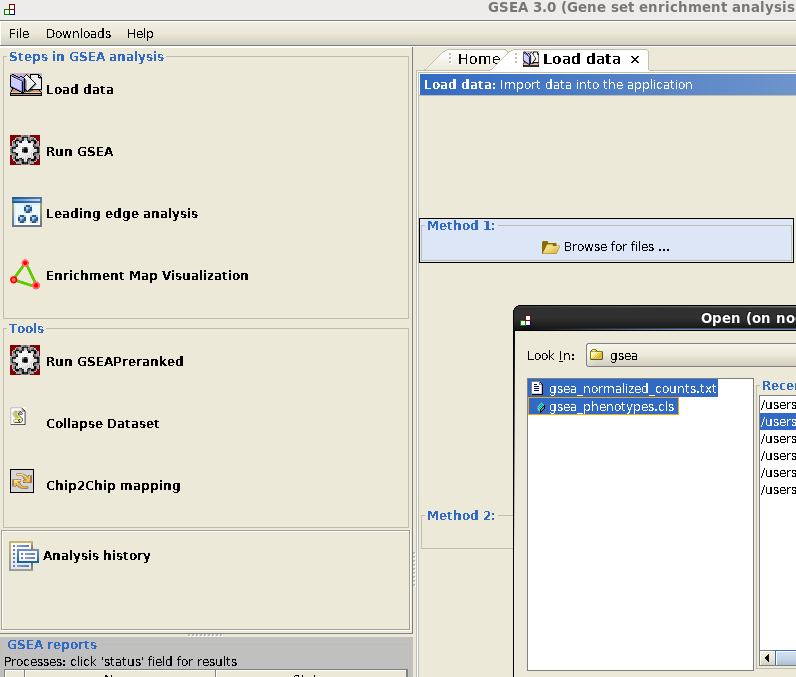
- Go to Run GSEA
- Results: index.html

- Enrichments results in html
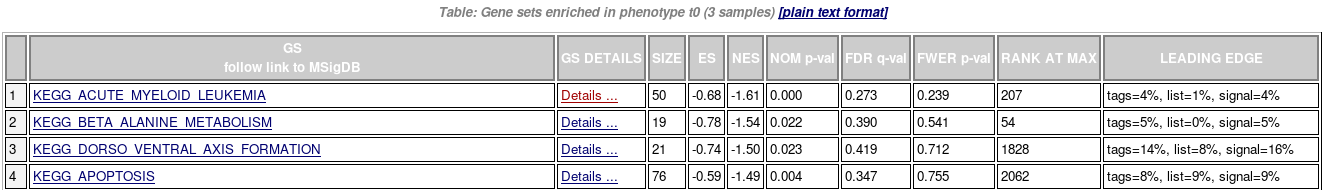
- Details for one gene set
- Summary of enrichment: phenotype, stats (Nominal p-value, FDR q-value, FWER p-value), enrichment scores (ES and Normalized ES)
- Enrichment plot
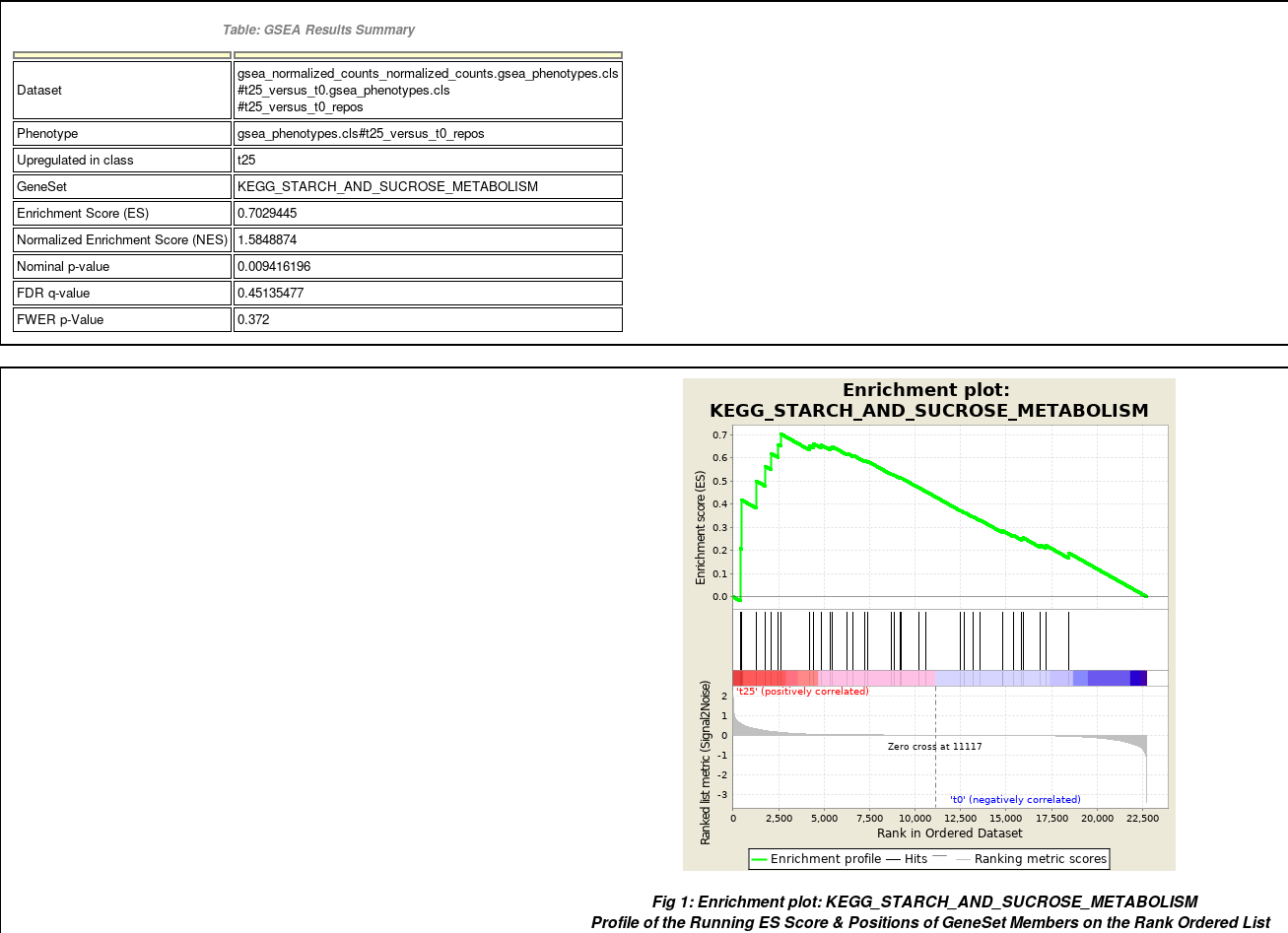
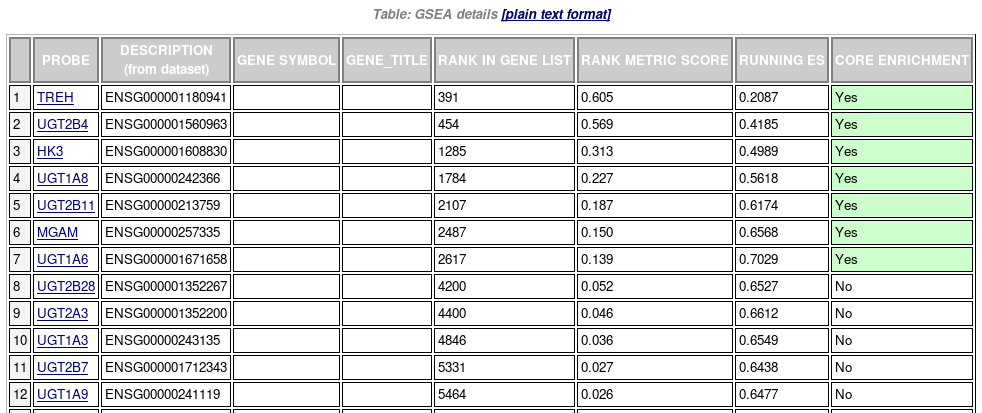
Table of genes: ranking, individual enrichment scores, core enrichment Yes/No.
From GSEA documentation, regarding core enrichment genes: “Genes with a Yes value in this column contribute to the leading-edge subset within the gene set. This is the subset of genes that contributes most to the enrichment result.”
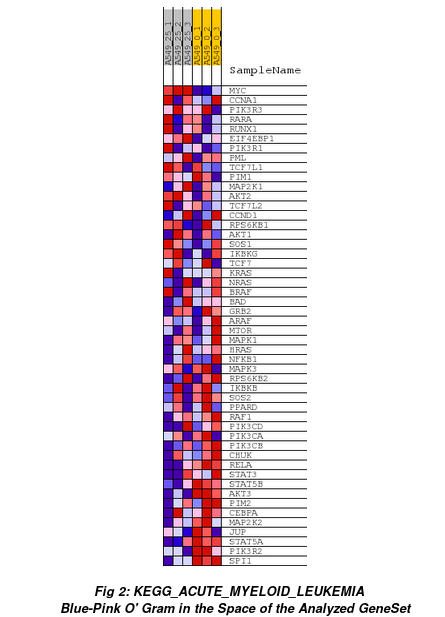
Heatmap of all genes from that gene set (ranked by GSEA) for each sample:
Suggestions for GSEA:
- Selection of pathways / gene sets: select the lowest FDR first.
- If you are looking for genes to validate on certain pathways:
- It is better if those genes belong to the core enrichment.
- It is also good to go back to the differential expression analysis table and make sure that their adjusted-value is low.
- You can also upload your own gene sets (for example a gene signature taken from a specific paper) to test against your list of genes, using one of the GSEA gene set database formats.

