Data used in this course
For the purpose of this course, we downloaded the following ENCODE data:
- Homo sapiens A549 treated with 100 nM dexamethasone for 0 minutes
- Homo sapiens A549 treated with 100 nM dexamethasone for 25 minutes
| Encode website |
|---|
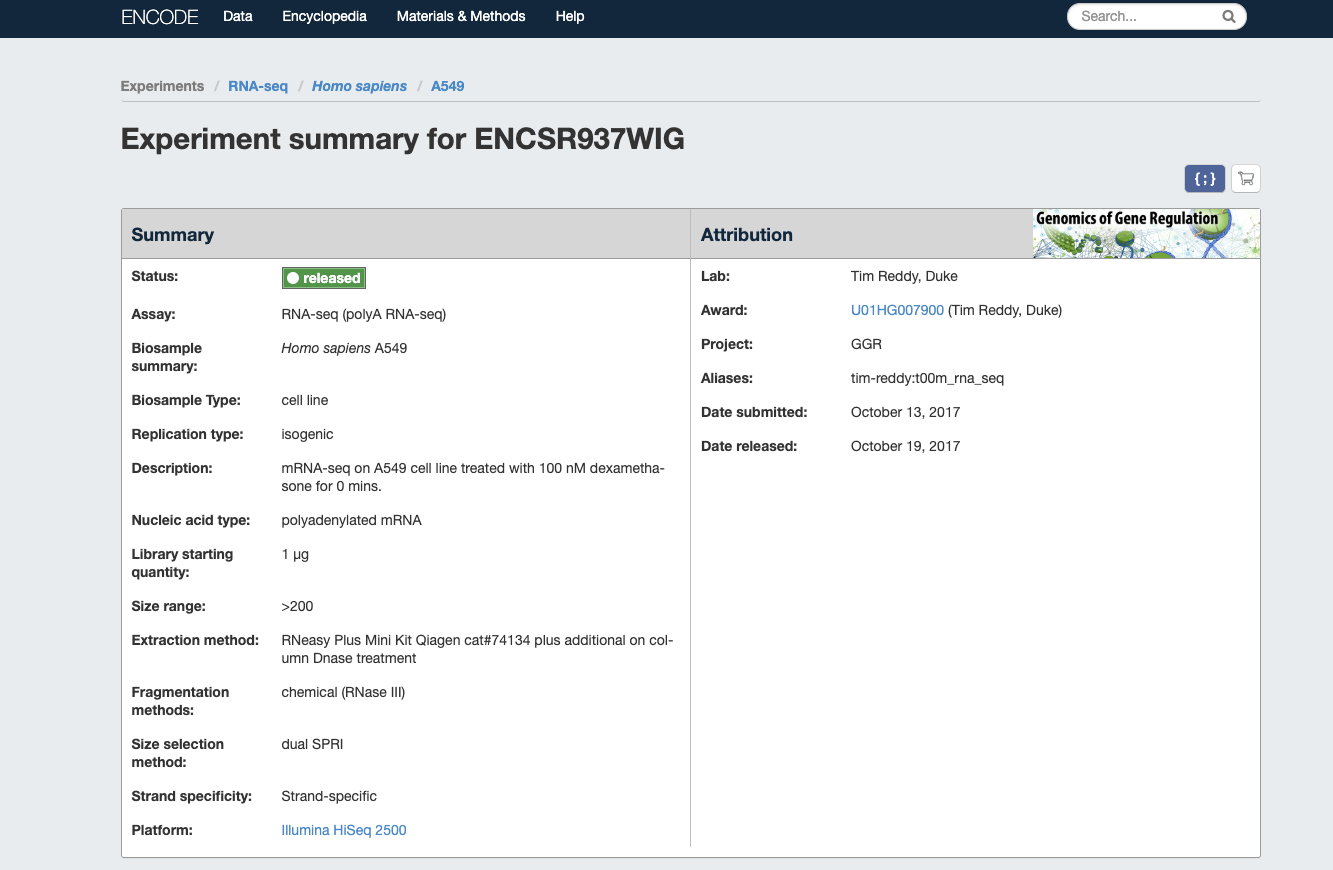 |
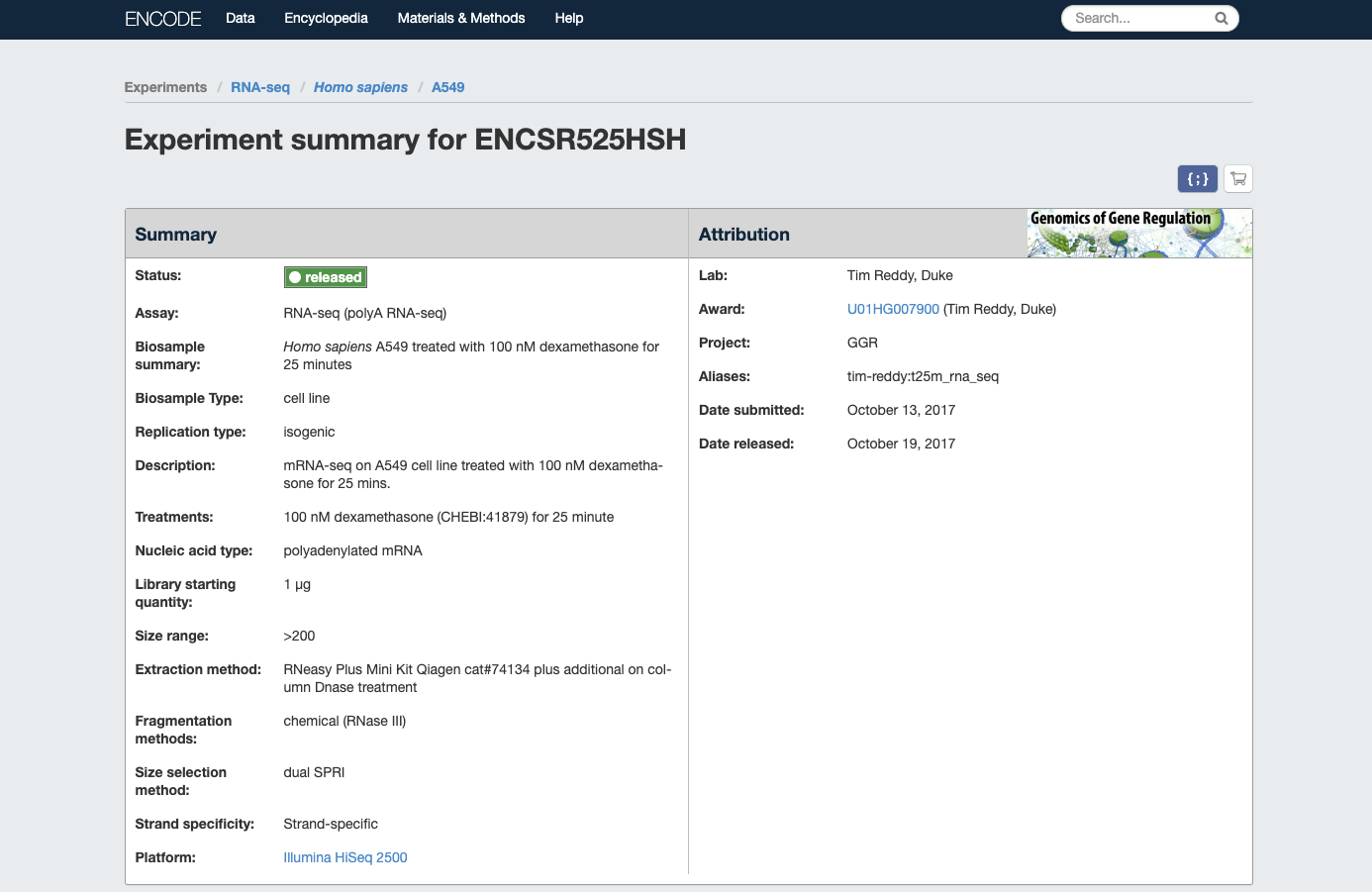 |
To download all fastq-files for this experiment takes a lot of time; therefore, to restrict the computation time of the read mapping, we selected reads that are mapped only to chromosome 10. Please run the following commands to obtain these files:
wget https://public-docs.crg.es/biocore/projects/training/RNAseq_2019/resources.tar
tar -vxf resources.tar
resources/
resources/A549_0_3chr10_1.fastq.gz
resources/A549_25_3chr10_2.fastq.gz
resources/A549_25_1chr10_1.fastq.gz
resources/A549_25_3chr10_1.fastq.gz
resources/A549_0_3chr10_2.fastq.gz
resources/A549_0_1chr10_1.fastq.gz
resources/A549_0_1chr10_2.fastq.gz
resources/A549_25_2chr10_1.fastq.gz
resources/A549_25_1chr10_2.fastq.gz
resources/A549_0_2chr10_1.fastq.gz
resources/A549_0_2chr10_2.fastq.gz
resources/A549_25_2chr10_2.fastq.gz
Let’s inspect these files, count the number of reads, and check the read length:
zcat resources/A549_25_3chr10_2.fastq.gz |more
@D00137:455:HLFL3BCXY:1:1111:7527:60273/2
GACAAACCCACAGCCAATATCATACTGAATGGGCAAAAACTGGAAGCATTC
+
ADDDDIIFHHIIIIIIIIIIHHHHIIIIHIIHHGIIIGIIIHHIIHHGHHH
@D00137:455:HLFL3BCXY:1:1111:3751:48736/2
CTATGGTGACCTGAACCACCTGGTGTCTGCTACCATGAGTGGGGTCACCAC
+
DDDDDIIIIIIIHIIHIIIIIIIIIIIIIHIIIIIIIIIIIIIHIIIIIIG
@D00137:455:HLFL3BCXY:2:1214:18935:42305/2
CTATGGTGACCTGAACCACCTGGTGTCTGCTACCATGAGTGGGGTCACCAC
+
DDDDDIIIHIIIIIIIIIIIIIIIIIIIIIIHIIIIIGHIIHIIIIIIIII
...
zcat resources/A549_25_3chr10_2.fastq.gz | awk '{num++} END{print num/4}'
2808343
....
zcat resources/A549_25_3chr10_2.fastq.gz | head -n 4 | tail -n 1 | awk '{print length($0)}'
51
EXERCISE
- Count the number of reads and check the read length for the Read 1 for the sample called A549_25_3chr10.
- Count the number of reads in all fastq files (use for-loop).

