Introduction: What is RNA-Seq?
| RNA molecule |
|---|
 |
| from Wikipedia |
RNA sequencing, aka RNA-Seq, is a technique that allows to detect and quantify RNA molecules within biological samples by using next-generation sequencing (NGS). This technology is used to analyze RNA by revealing
- RNA/gene/transcript expression;
- alternatively spliced transcripts;
- gene fusion and SNPs;
- post-translational modification.
Other technologies for assessing RNA expression are Northern Blot, real-time PCR and hybridization-based microarrays [3].
RNA-Seq can be performed using:
- total RNA (in this case the content of ribosomal RNA is about 80%);
- rRNA depleted RNA (after removing ribosomal RNA);
- mRNA transcripts (by performing polyA enrichment of RNA);
- small RNA, such as miRNA, tRNA (by selecting the size of RNA molecules; e.g., < 100 nt);
- RNA molecules transcribed at a specific time (ribosomal profiling);
- specific RNA molecules (via hybridization with probes complementary to desired transcripts).
Depending on the technology and the protocol, RNA-Seq can produce:
- single-end short reads (50-450 nt), which are useful for gene expression quantification (mainly Illumina, but also Ion Torrent and BGISEQ);
- paired-end reads (2 x 50-250 nt), which are useful for detecting splicing events and refinement of transcriptome annotation;
- single long reads (PACBio or Nanopore), which are used for the de novo identification of new transcripts and improving transcriptome assembly.
mRNA-Seq protocol (Illumina)
| mRNA-Seq protocol |
|---|
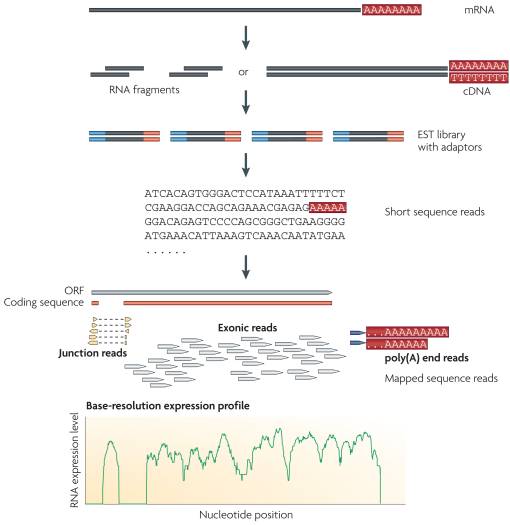 |
| from Wang et al 2009 |

