Pre-processing of sequencing reads
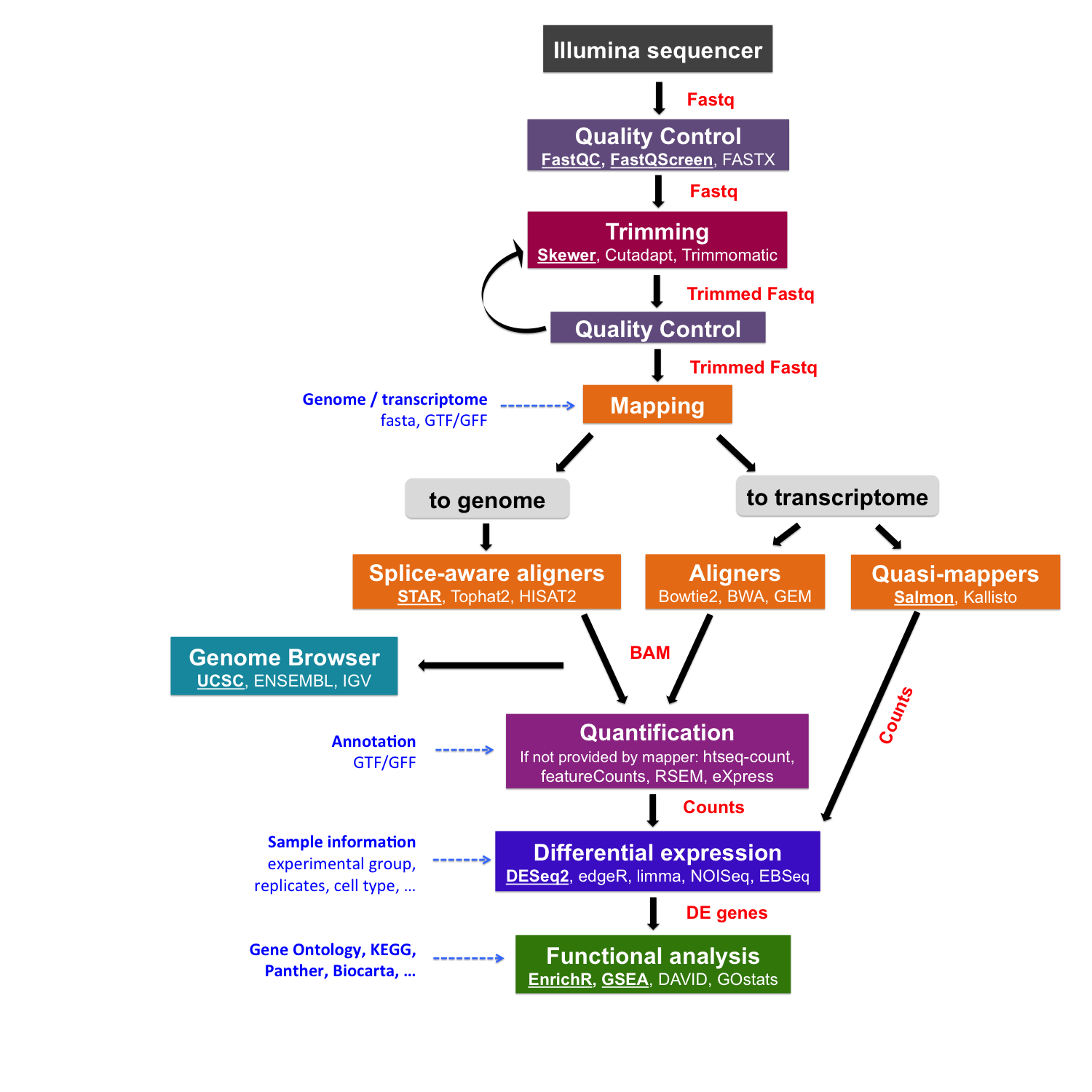
The raw data obtained from the sequencing machine need to be pre-processed.
Pre-processing can include the quality control of initial reads and read trimming that includes removing adapter sequences, filtering out low quality reads and trimming reads off low quality base pairs.
Quality control of sequencing reads
FastQC
To assess the quality of sequencing data, we will use the programs FastQC and Fastq Screen.
FastQC calculates statistics about the composition and quality of raw sequences, while Fastq Screen looks for possible contaminations.
# Go to the "quality_control" folder
cd ~/rnaseq_course/quality_control
# Run FastQC for one sample
$RUN fastqc ~/rnaseq_course/raw_data/fastq_chr6/SRR3091420_1_chr6.fastq.gz -o .
# Run for all samples
$RUN fastqc ~/rnaseq_course/raw_data/fastq_chr6/*fastq.gz -o .
The output files are a .zip archive and an .html file
We can display the results (.html file) with an Internet browser; e.g. Firefox:
firefox SRR3091420_1_chr6_fastqc.html &
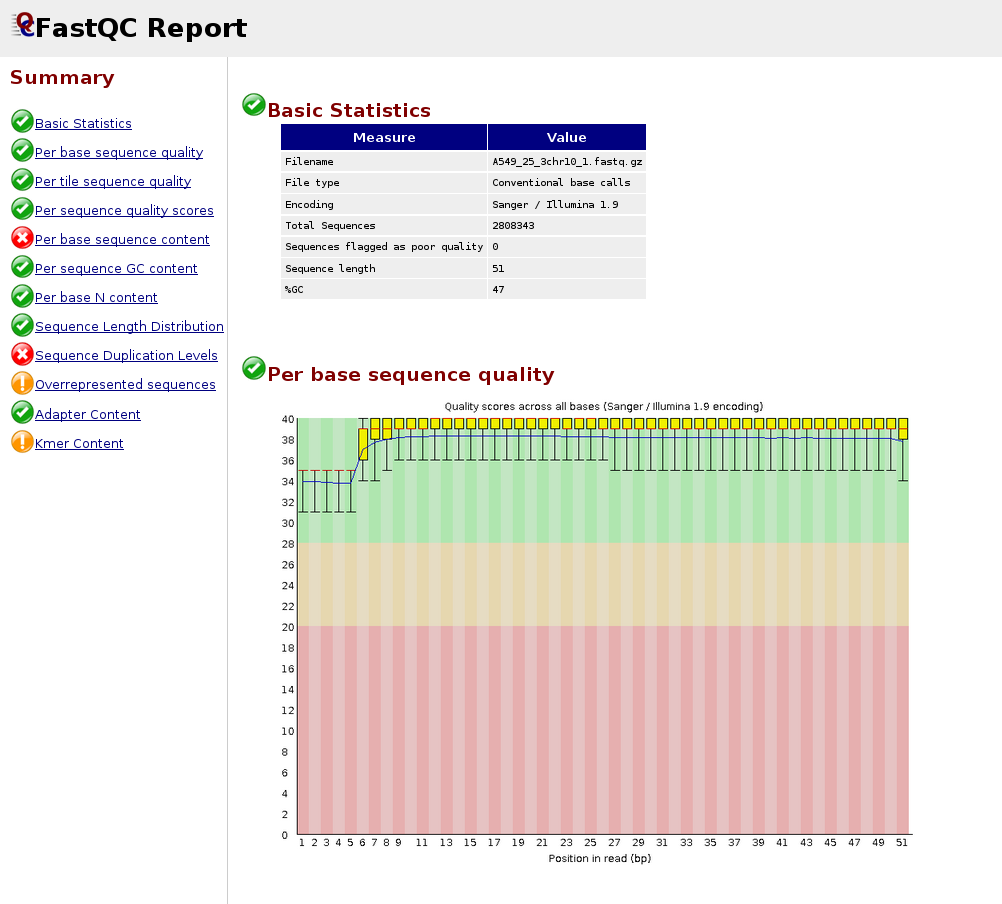
Below is an example of a poor quality dataset. As you can see, the average quality drops dramatically towards the 3’-end.
You can extract the files from the .zip archive:
# extract
unzip SRR3091420_1_chr6_fastqc.zip
# remove remaining .zip file
rm SRR3091420_1_chr6_fastqc.zip
# display content of directory
ls SRR3091420_1_chr6_fastqc
File fastqc_data.txt contains the results in text format, for easier parsing of the results:
less SRR3091420_1_chr6_fastqc/fastqc_data.txt
FastQ Screen
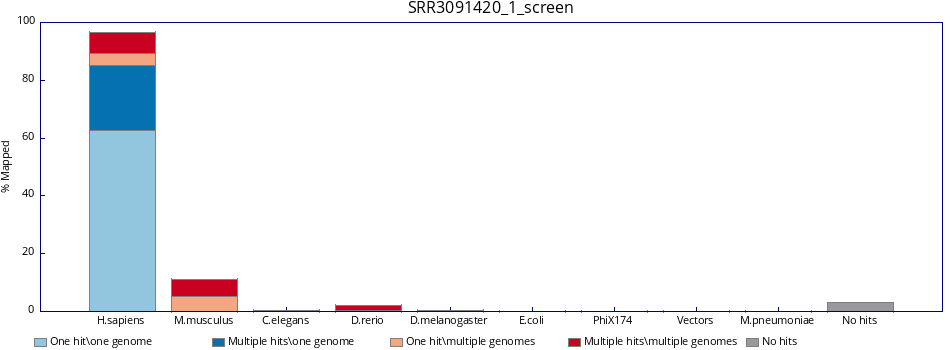
FastQ Screen is a tool that allows to screen libraries for potential contaminations.
It requires to download genome indices (data bases) from a variety of organisms, a lot of which can be downloaded by default.
Note that fastq_screen can be found in the singularity image, but not the data bases!
WARNING: do not run the following command in class: it will take too much time and resources!
# Download default data bases
$RUN fastq_screen --get_genomes
The latter commands download 14 indexes from Bowtie2 mapper (from model organisms or known contaminants):
- Arabidopsis thaliana
- Drosophila melanogaster
- Escherichia coli
- Homo sapiens
- Mus musculus
- Rattus norvegicus
- Caenorhabditis elegans
- Saccharomyces cerevisiae
- Lambda
- Mitochondria
- PhiX
- Adapters
- Vectors
- rRNA
Upon download, the FastQ_Screen_Genomes folder is created, containing all indexes.
The file fastq_screen.conf will be also downloaded in this folder: in order to use the tool, you will have to modify the fastq_screen.conf by providing the full path to the Bowtie2 executable (/usr/local/bin/bowtie2 if you use our singularity image) and full paths to the downloaded folders with genome index files.
Here we show where to change the executables:
# This is a configuration file for fastq_screen
###########
## Bowtie #
###########
## If the bowtie binary is not in your PATH then you can
## set this value to tell the program where to find it.
## Uncomment the line below and set the appropriate location
##
#BOWTIE /usr/local/bin/bowtie/bowtie
BOWTIE2 /usr/local/bin/bowtie2
...
FastQ Screen runs checks on a random subset of 100,000 reads (that can be changed using option –subset).
You can execute FastQ Screen this way:
$RUN fastq_screen --conf FastQ_Screen_Genomes/fastq_screen.conf \
~/rnaseq_course/raw_data/SRR3091420_1.fastq.gz \
--outdir ~/rnaseq_course/quality_control/
Below is an example of the FastQ Screen results for SRR3091420_1.fastq.gz.
cd ~/rnaseq_course/quality_control/
# get files from fastq_screen run
wget https://public-docs.crg.es/biocore/projects/training/PHINDaccess2020/SRR3091420_1_fastq_screen.tar.gz
# extract
tar -zvxf SRR3091420_1_fastq_screen.tar.gz
firefox SRR3091420_1_screen.html
Initial processing of sequencing reads
Before mapping reads to the genome/transcriptome or performing a de novo assembly, the reads have to be pre-processed, if needed, as follows:
- Demultiplex by index or barcode (usually done in the sequencing facility)
- Remove/Trim adapter sequences
- Trim reads by quality
- Discard reads by quality/ambiguity
- Filter reads by k-mer coverage (recommended for the de novo assembly)
- Normalize k-mer coverage (recommended for the de novo assembly)
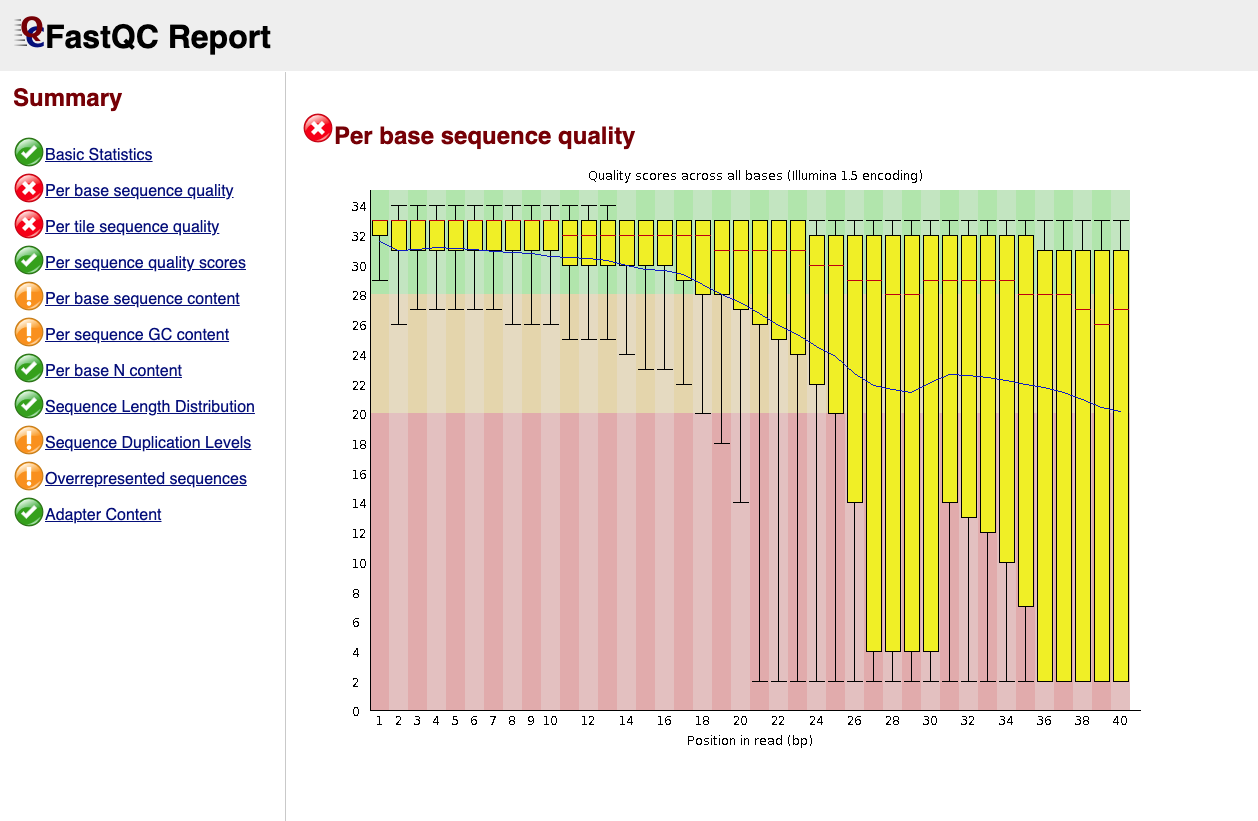
As shown before, both the presence of low quality reads and adapters are reported in the fastqc output.
Adapters are usually expected in small RNA-Seq because the molecules are typically shorter than the reads, and that makes an adapter to be present at 3’-end.
Below is an example of the FastQC report for a small RNA-seq sample:
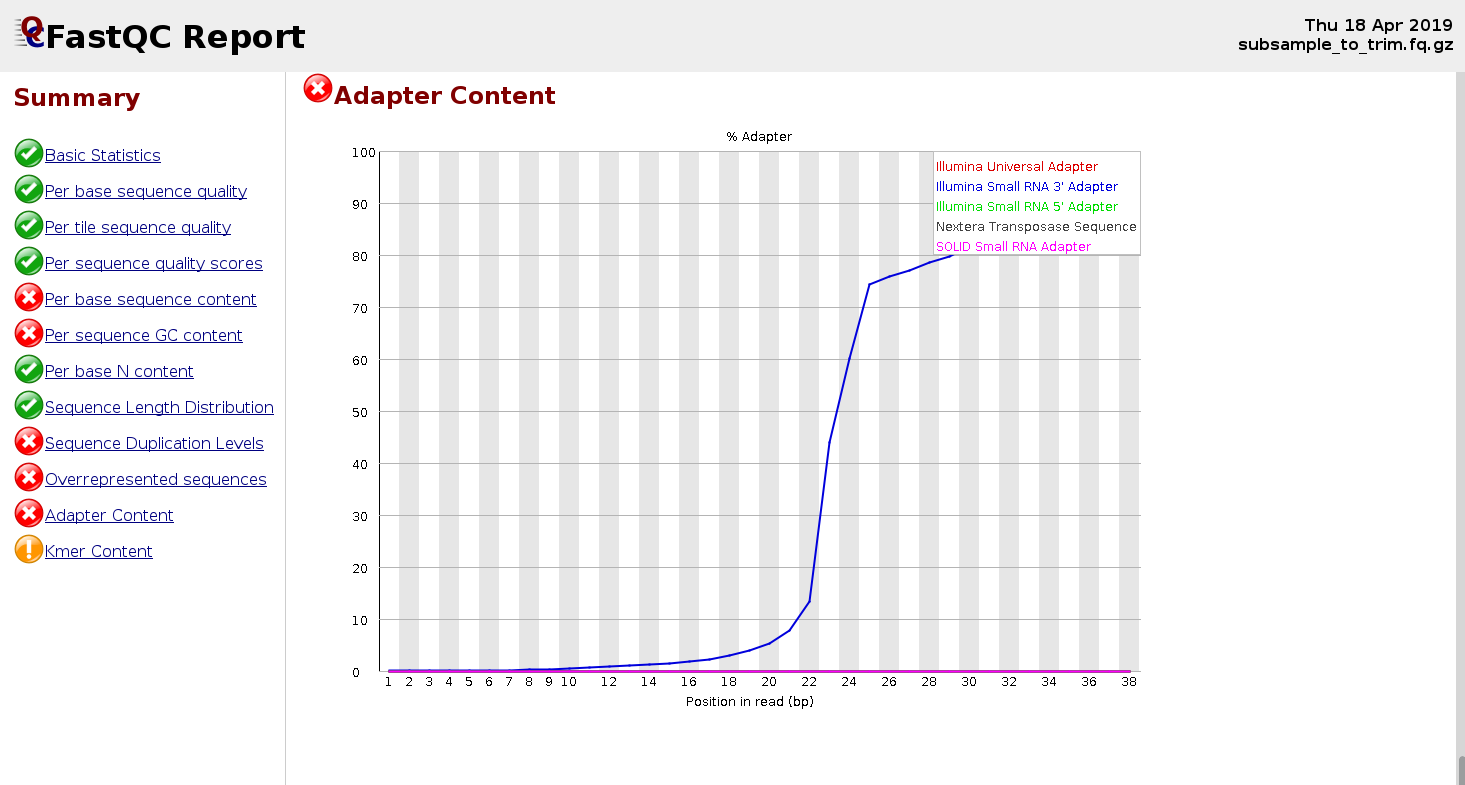
Trimming
There are many tools for trimming reads and removing adapters, such as Trim Galore!, Trimmomatic, Cutadapt, skewer, AlienTrimmer, BBDuk, and the most recent SOAPnuke and fastp.
Let’s use skewer to trim the Illumina 3’ adapter.
cd ~/rnaseq_course/trimming
$RUN skewer ~/rnaseq_course/raw_data/fastq_chr6/SRR3091420_1_chr6.fastq.gz \
-x TGGAATTCTCGGGTGCCAAGG \
-o SRR3091420_1_chr6 \
-z
.--. .-.
: .--': :.-.
`. `. : `'.' .--. .-..-..-. .--. .--.
_`, :: . `.' '_.': `; `; :' '_.': ..'
`.__.':_;:_;`.__.'`.__.__.'`.__.':_;
skewer v0.2.2 [April 4, 2016]
Parameters used:
-- 3' end adapter sequence (-x): TGGAATTCTCGGGTGCCAAGG
-- maximum error ratio allowed (-r): 0.100
-- maximum indel error ratio allowed (-d): 0.030
-- minimum read length allowed after trimming (-l): 18
-- file format (-f): Solexa/Illumina 1.3+/Illumina 1.5+ FASTQ (auto detected)
-- minimum overlap length for adapter detection (-k): 3
Tue Feb 4 11:00:56 2020 >> started
|=================================================>| (100.00%)
Tue Feb 4 11:01:40 2020 >> done (43.669s)
12157169 reads processed; of these:
0 ( 0.00%) short reads filtered out after trimming by size control
0 ( 0.00%) empty reads filtered out after trimming by size control
12157169 (100.00%) reads available; of these:
480367 ( 3.95%) trimmed reads available after processing
11676802 (96.05%) untrimmed reads available after processing
log has been saved to "SRR3091420_1-trimmed.log".
We can look at the read distribution after the trimming of the adapter by inspecting the log-file or re-launching FastQC.
cd ~/rnaseq_course/quality_control/
$RUN fastqc ~/rnaseq_course/trimming/SRR3091420_1_chr6-trimmed.fastq.gz -o .
firefox SRR3091420_1_chr6-trimmed_fastqc.html &
Example of a FastQC report for a trimmed small-RNA sample:
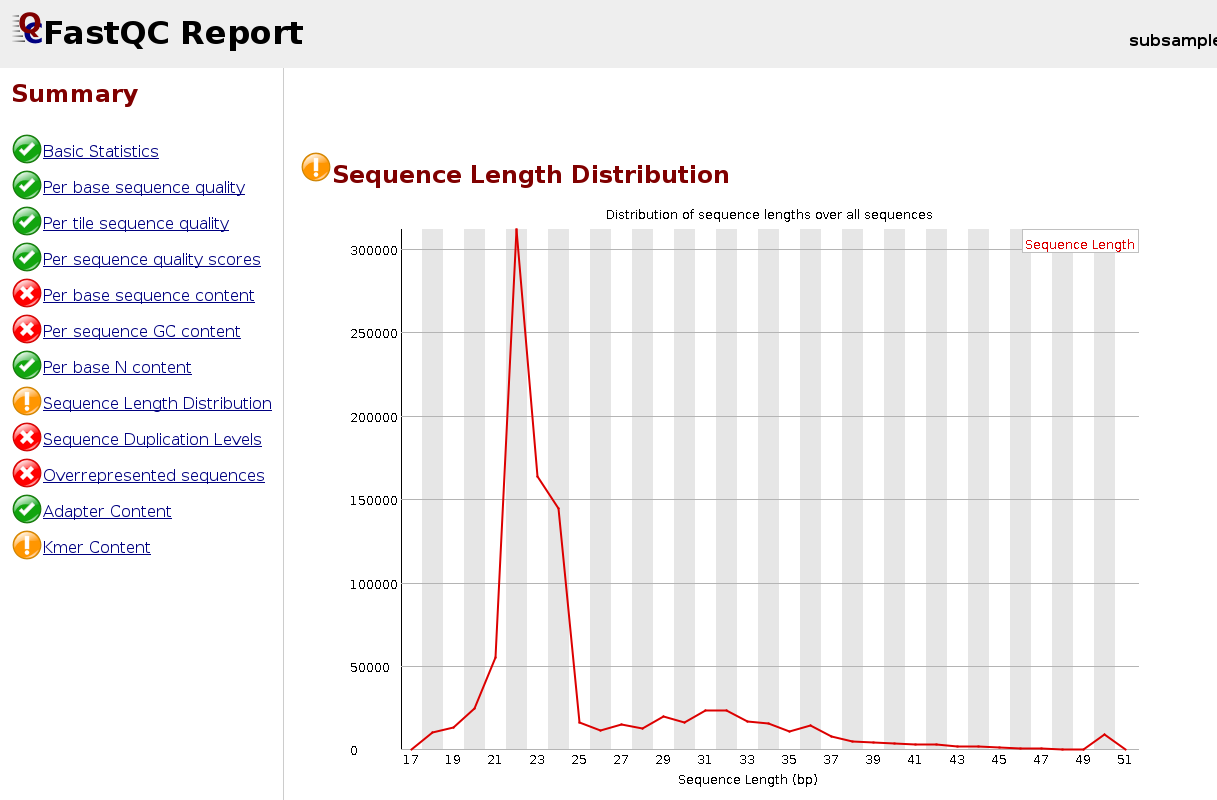
Now run skewer for all samples
for fastq in ~/rnaseq_course/raw_data/fastq_chr6/*fastq.gz
do echo $fastq
$RUN skewer $fastq \
-x TGGAATTCTCGGGTGCCAAGG \
-o $(basename $fastq .fastq.gz) \
-z
done
EXERCISE
Let’s explore the tool skewer in more detail: use “skewer –help” for a description of the parameters.
- Which parameter indicates the minimum read length allowed after trimming? What is its default value?
- Which parameter indicates the threshold on the average read quality to be filtered out?
- Using skewer, filter out reads in “SRR3091420_1-trimmed.fastq” that have average quality below 30 and trim them on 3’-end until the base quality has reached 30. How many reads were filtered out and how many remain?

