Mapping using STAR
For the STAR running options, see STAR Manual.
Building the STAR index
To make an index for STAR, we need both the genome sequence in FASTA format and the annotation in GTF format.
As STAR is very resource consuming, we will create an index for chromosome 6 only (and hope that it will work!). We already downloaded the FASTA and GTF files (in ~/rnaseq_course/reference_genome) needed for the indexing.
However, STAR requires unzipped .fa and .gtf files. We need to unzip them.
# go to reference_genome folder
cd ~/rnaseq_course/reference_genome/reference_chr6
# unzip files (keep original zipped file)
zcat Homo_sapiens.GRCh38.88.chr6.gtf.gz > Homo_sapiens.GRCh38.88.chr6.gtf
zcat Homo_sapiens.GRCh38.dna.chrom6.fa.gz > Homo_sapiens.GRCh38.dna.chrom6.fa
Q. How much (in percentage) disk space is saved when those two files are kept zipped vs unzipped?
Once the index is built, do not forget to remove those unzipped files!
To index the genome with STAR for RNA-seq analysis, the sjdbOverhang option needs to be specified for detecting possible splicing sites:
- It usually equals the minimum read size minus 1; it tells STAR what is the maximum possible stretch of sequence that can be found on one side of a spicing site.
- In our case, since the read size is 49 bases, we can accept maximum 48 bases on one side and one base on the other of a splicing site; that is, to set up this parameter to 48.
- This also means that for every different read-length to be aligned a new STAR index needs to be generated. Otherwise a drop in aligned reads can be experienced.
- –runThreadN allows you to parallelize the job.
NOTE that for small genomes, parameter –genomeSAindexNbases (default 14) should be scaled down as: min(14, log2(GenomeLength)/2 - 1). Here: min(14, log2(170805979/2)-1) =~ 12.6
Building the STAR index (option –runMode genomeGenerate):
# go to mapping folder
cd ~/rnaseq_course/mapping
# create sub-folder where index will be generated
mkdir index_star_chr6
# create the index and store it in ~/rnaseq_course/mapping/index_star_chr6
$RUN STAR --runMode genomeGenerate --genomeDir index_star_chr6 \
--genomeFastaFiles ~/rnaseq_course/reference_genome/reference_chr6/Homo_sapiens.GRCh38.dna.chrom6.fa \
--sjdbGTFfile ~/rnaseq_course/reference_genome/reference_chr6/Homo_sapiens.GRCh38.88.chr6.gtf \
--sjdbOverhang 48 \
--genomeSAindexNbases 12.6 \
--outFileNamePrefix Hsapiens_chr6 \
--runThreadN 4
- –genomeSAindexNbases: default 14. If genome is small, should be scaled down as: min(14, log2(GenomeLength)/2 - 1). Here: min(14, log2(170805979/2)-1) =~ 12.6
Aligning reads to the genome (and counting them at the same time!)
To use STAR for the read alignment (default –runMode option), we have to specify the following options:
- the index directory (–genomeDir)
- the read files (–readFilesIn)
- if reads are compressed or not (–readFilesCommand)
The following parameters are optional:
- –outSAMtype: type of output. Default is BAM Unsorted; STAR outputs unsorted Aligned.out.bam file(s). “The paired ends of an alignment are always adjacent, and multiple alignments of a read are adjacent as well. This ”unsorted” file cannot be directly used with downstream software such as HTseq, without the need of name sorting.” We therefore prefer the option BAM SortedByCoordinate
- –outFileNamePrefix: the path for the output directory and prefix of all output files. By default, this parameter is ./, i.e. all output files are written in the current directory.
- –quantMode. With the –quantMode GeneCounts option set, STAR will count the number of reads per gene while mapping. A read is counted if it overlaps (1nt or more) one and only one gene. In case of mapping paired-end data, both ends are checked for overlaps. The counts coincide with those produced by the htseq-count tool with default parameters. This option requires annotations (GTF or GFF with –-sjdbGTFfile option) used at the genome generation step, or at the mapping step. (from STAR Manual)
We can try and launch the mapping for one file:
# go to mapping folder
cd ~/rnaseq_course/mapping
# create sub-folder where we will store the alignments
mkdir alignments_STAR
$RUN STAR --genomeDir index_star_chr6 \
--readFilesIn ~/rnaseq_course/trimming/SRR3091420_1_chr6-trimmed.fastq.gz \
--readFilesCommand zcat \
--outSAMtype BAM SortedByCoordinate \
--quantMode GeneCounts \
--outFileNamePrefix alignments_STAR/SRR3091420_1_chr6 \
--runThreadN 4
If this was successful and not too slow and resource consuming, you can do it for all samples, in a loop:
for fastq in ~/rnaseq_course/trimming/*-trimmed.fastq.gz
do echo $fastq
$RUN STAR --genomeDir index_star_chr6 \
--readFilesIn $fastq \
--readFilesCommand zcat \
--outSAMtype BAM SortedByCoordinate \
--quantMode GeneCounts \
--outFileNamePrefix alignments_STAR/$(basename $fastq -trimmed.fastq.gz)
done
BACKUP !!
If it was indeed too resource consuming, you can download the aligned files in BAM format from:
cd ~/rnaseq_course/mapping
# get archive
wget https://public-docs.crg.es/biocore/projects/training/PHINDaccess2020/bam_chr6.tar.gz
# extract
tar -xvzf bam_chr6.tar.gz
Let’s explore the output directory “alignments” (or “bam_chr6”, if we used the backup folder).
ls -lh alignments
ls -lh bam_chr6
Read counts
STAR outputs read counts per gene into PREFIXReadsPerGene.out.tab file with 4 columns which correspond to different strandedness options:
| column 1 | gene ID |
|---|---|
| column 2 | counts for unstranded RNA-seq |
| column 3 | counts for the 1st read strand aligned with RNA (htseq-count option -s yes) |
| column 4 | counts for the 2nd read strand aligned with RNA (htseq-count option -s reverse) |
Let’s see what the ReadsPerGene.out.tab file looks like for sample SRR3091420_1_chr6:
head alignments/SRR3091420_1_chr6ReadsPerGene.out.tab
head bam_chr6/SRR3091420_1_chr6-trimmedReadsPerGene.out.tab
| gene id | read counts per gene (unstranded) | read counts per gene (read 1) | read counts per gene (read 2) |
| N_unmapped | 1589 | 1589 | 1589 |
| N_multimapping | 45100 | 45100 | 45100 |
| N_noFeature | 33480 | 393427 | 413797 |
| N_ambiguous | 29733 | 7977 | 7339 |
| ENSG00000271530 | 0 | 0 | 0 |
| ENSG00000220212 | 0 | 0 | 0 |
| ENSG00000170590 | 2 | 2 | 0 |
Select the output according to the strandedness of your data.
Note, if you have stranded data and choose one of the columns 3 or 4, the other column (4 or 3) will give you the count of antisense reads.
For example, in the stranded protocol shown in “Library preparation”, Read 1 is mapped to the antisense strand (this is also true for single-end reads), while Read 2, to the sense strand.
Which protocol, stranded or unstranded, was used for this RNA-seq data?
We can count the number of reads mapped to each strand by using a simple awk script:
grep -v "N_" alignments/SRR3091420_1_chr6-trimmedReadsPerGene.out.tab | awk '{unst+=$2;forw+=$3;rev+=$4}END{print unst,forw,rev}'
# OR
grep -v "N_" bam_chr6/SRR3091420_1_chr6-trimmedReadsPerGene.out.tab | awk '{unst+=$2;forw+=$3;rev+=$4}END{print unst,forw,rev}'
# 725267 387076 367344
We see that 387,076 Reads 1 (forward) were mapped to known genes and 367,344 Reads 2 (reverse) were mapped to known genes.
These numbers are very similar, which indicates that the protocol used for this mRNA-Seq experiment is unstranded.
If the protocol used was stranded, there would be a strong imbalance between number of reads mapped to known genes in forward versus reverse strands.
BAM/SAM/CRAM format
The BAM format is a compressed version of the SAM format (which is a plain text) and cannot thus being seen as a text. To explore the BAM file, we have to convert it to the SAM format by using samtools. Note that we use the parameter -h to show also the header that is hidden by default.
$RUN samtools view -h bam_chr6/SRR3091420_1_chr6-trimmedAligned.sortedByCoord.out.bam | head -n 10
@HD VN:1.4 SO:coordinate
@SQ SN:6 LN:170805979
@PG ID:STAR PN:STAR VN:STAR_2.5.3a CL:STAR --genomeDir index_chr6 --readFilesIn ../RNAseq/output_nextflow/Alignments/selection_chr6/SRR3091420_1_chr6.fastq.gz --readFilesCommand zcat --outFileNamePrefix alignments/SRR3091420_1_chr6 --outSAMtype BAM SortedByCoordinate --quantMode GeneCounts
@CO user command line: STAR --genomeDir index_chr6 --readFilesIn ../RNAseq/output_nextflow/Alignments/selection_chr6/SRR3091420_1_chr6.fastq.gz --readFilesCommand zcat --outSAMtype BAM SortedByCoordinate --quantMode GeneCounts --outFileNamePrefix alignments/SRR3091420_1_chr6
10416098 0 6 113167 255 49M * 0 0 GGGAAAAGTACAAATTCAACATGTAATTGTATAGTAATCCATATAAAAA bbbeeeeecggggiiiiiiiiiihhhiiighhiihhhhigiiiiiiiih NH:i:1 HI:i:1 AS:i:48 nM:i:0
8553177 272 6 119288 3 1S48M * 0 0 TGAAATCCAGTGGGACAGTCAAATCTTAAAGCTCCAAAATGATCTCCTT hiiiiiiiiiiiiigiiiiiiiihiihiiiiihhiigggggeeeeebbb NH:i:2 HI:i:2 AS:i:47 nM:i:0
4630026 272 6 128432 3 49M * 0 0 AGCACTAACCATTGTAGCATGCCAATATACTCAAAATTCAATGAAATTC hfgehhggiihhhiiihhiihhhhffffghdihhiifggggeeeeebbb NH:i:2 HI:i:2 AS:i:48 nM:i:0
4630026 272 6 128432 3 49M * 0 0 AGCACTAACCATTGTAGCATGCCAATATACTCAAAATTCAATGAAATTC hfgehhggiihhhiiihhiihhhhffffghdihhiifggggeeeeebbb NH:i:2 HI:i:2 AS:i:48 nM:i:0
10689795 0 6 135934 255 49M * 0 0 AAGGCTGCAATGAGCTGTGATCGCACCACCGCACCCAAGCCTGGGTGGT bbbeeeeeggggfiiiighiiiiiiiiiiiiiihiihfhhiiiii_ega NH:i:1 HI:i:1 AS:i:44 nM:i:2
10416101 0 6 136561 255 49M * 0 0 CCCAACGTTTAGACTACACAATGAGTTAAGAACGACAAAAATAAGCTCA ___ecccceeeeghhhhhhhhgfgiihfhhhhfhffffgghhhidfffh NH:i:1 HI:i:1 AS:i:48 nM:i:0
The first part indicated by the first character @ in each row is the header:
| Symbol | ||
|---|---|---|
| @HD header line | VN:1.4 version of the SAM format | SO:coordinate sorting order |
| @SQ reference sequence dictionary | SN:6 sequence name | LN:170805979 sequence length |
| @PG program used | ID:STAR PN:STAR VN:2.5.3a version | CL:STAR –genomeDir index_chr6 –readFilesIn ../RNAseq/output_nextflow/Alignments/selection_chr6/SRR3091420_1_chr6.fastq.gz –readFilesCommand zcat –outFileNamePrefix alignments/SRR3091420_1_chr6 –outSAMtype BAM SortedByCoordinate –quantMode GeneCounts command line |
| @CO One-line text comment | user command line: STAR –genomeDir index_chr6 –readFilesIn ../RNAseq/output_nextflow/Alignments/selection_chr6/SRR3091420_1_chr6.fastq.gz –readFilesCommand zcat –outSAMtype BAM SortedByCoordinate –quantMode GeneCounts –outFileNamePrefix alignments/SRR3091420_1_chr6 |
The rest is a read alignment.
| Field | Value |
|---|---|
| Query name | 8553177 |
| FLAG | 272 * |
| Reference name | 6 |
| Leftmost mapping position (1-based) | 119288 |
| Mapping quality | 3 (p=0.5) |
| CIGAR string | 1S48M * |
| Reference sequence name of the primary alignment of the mate | * = no mate (= same chromosome) |
| Position of the primary alignment of the mate | 0 |
| observed fragment length | 0 |
| Sequence | TGAAATCCAGTGGGACAGTCAAATCTTAAAGCTCCAAAATGATCTCCTT |
| Quality | hiiiiiiiiiiiiigiiiiiiiihiihiiiiihhiigggggeeeeebbb |
* FLAG 272 means that the read is non paired, and that it maps on the reverse strand.
CIGAR string 1S48M means that 1 base was soft clip (S) and 48 bases were mapped to the reference (M). N would correspond to bases unmapped.
You can use this website for the translation of SAM FLAG values and this one for interpreting CIGAR strings.
Extra fields are often present and differ between aligners https://samtools.github.io/hts-specs/SAMtags.pdf. In our case we have:
| Field | Meaning |
|---|---|
| NH:i:2 | number of mapping to the reference |
| HI:i:2 | which alignment is the reported one (in this case is the second one) |
| AS:i:74 | Alignment score calculate by the aligner |
| nM:i:9 | number of difference with the reference* |
* Note that historically this has been ill-defined and both data and tools exist that disagree with this definition.
Let’s convert BAM to SAM:
$RUN samtools view -h bam_chr6/SRR3091420_1_chr6-trimmedAligned.sortedByCoord.out.bam > bam_chr6/SRR3091420_1_chr6Aligned.sortedByCoord.out.sam
You can see that the SAM file is 5 times larger than the BAM file.
Yet, the more efficient way to store the alignment is to use the CRAM format. CRAM is fully compatible with BAM, and main repositories, such as GEO and SRA, accept alignments in the CRAM format. UCSC Genome Browser can visualize both BAM and CRAM files. It is now a widly recommended format for storing alignments.
To convert BAM to CRAM, we have to provide unzipped and indexed version of the genome.
$RUN samtools faidx ~/rnaseq_course/reference_genome/reference_chr6/Homo_sapiens.GRCh38.dna.chrom6.fa
$RUN samtools view -C bam_chr6/SRR3091420_1_chr6-trimmedAligned.sortedByCoord.out.bam -T ~/rnaseq_course/reference_genome/reference_chr6/Homo_sapiens.GRCh38.dna.chrom6.fa > bam_chr6/SRR3091420_1_chr6Aligned.sortedByCoord.out.cram
You can see that a .cram file is twice as small as a .bam file.
Let’s remove the .sam file:
rm bam_chr6/*.sam
Alignment QC
The quality of the resulting alignment can be checked using the tool QualiMap. To run QualiMap, we specify the kind of analysis (rnaseq), the gtf file, the strandness of the library (-p unstranded).
*Note that if the library was paired-end, you would add the -pe option.
**IMPORTANT: before running QualiMap ensure enough disk space for a temporary directory ./tmp that the program is required, running the following command:
export _JAVA_OPTIONS="-Djava.io.tmpdir=./tmp -Xmx6G"
cd ~/rnaseq_course/mapping
# create folder
mkdir qc_qualimap
# run qualimap
$RUN qualimap rnaseq -bam bam_chr6/SRR3091420_1_chr6-trimmedAligned.sortedByCoord.out.bam \
-gtf ~/rnaseq_course/reference_genome/reference_chr6/Homo_sapiens.GRCh38.88.chr6.gtf \
-outdir qc_qualimap \
-p non-strand-specific
We can check the final report in a browser:
firefox qc_qualimap/qualimapReport.html
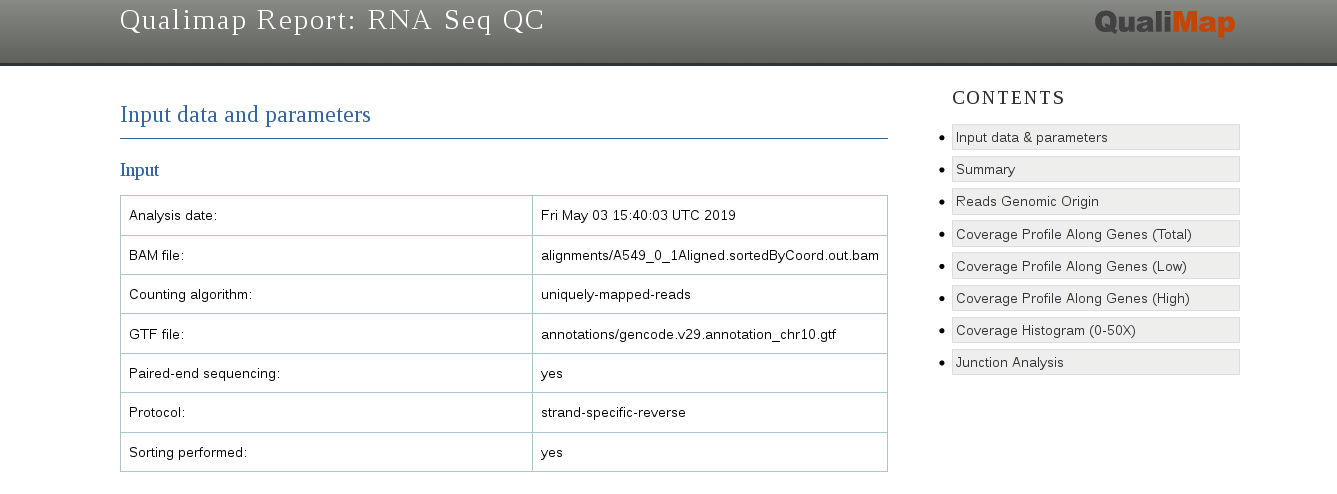
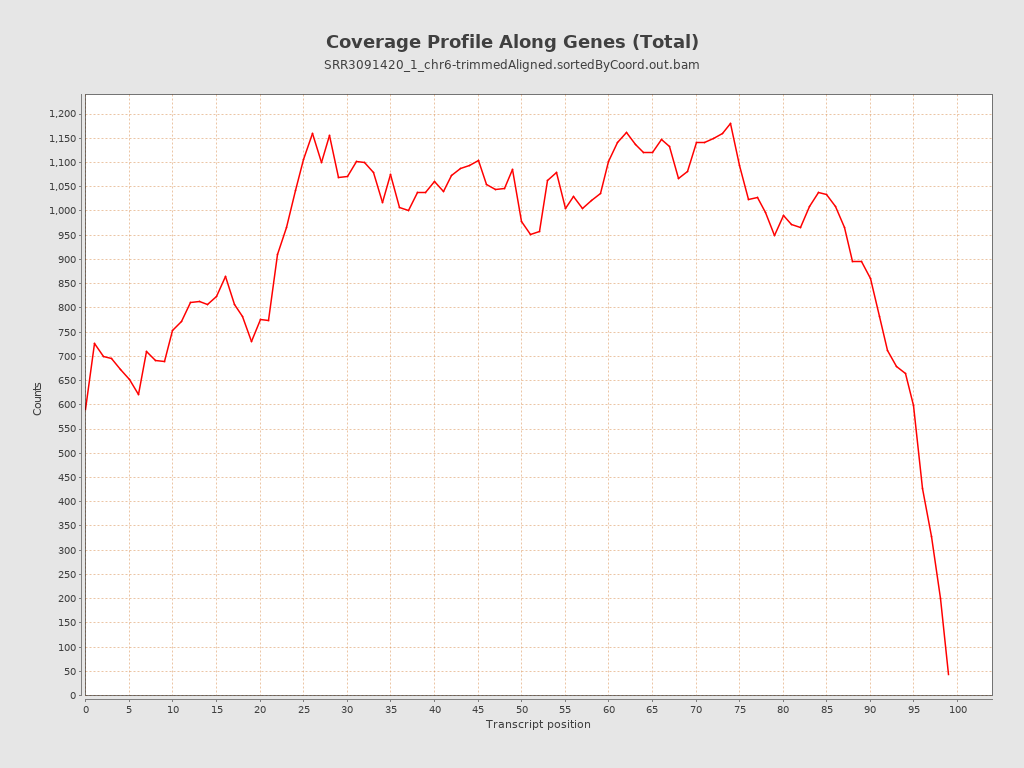
The report gives a lot of useful information, such as the total number of mapped reads, the amount of reads mapped to exons, introns or intergenic regions, and the bias towards one of the ends of mRNA (that can give information about RNA integrity or a protocol used).
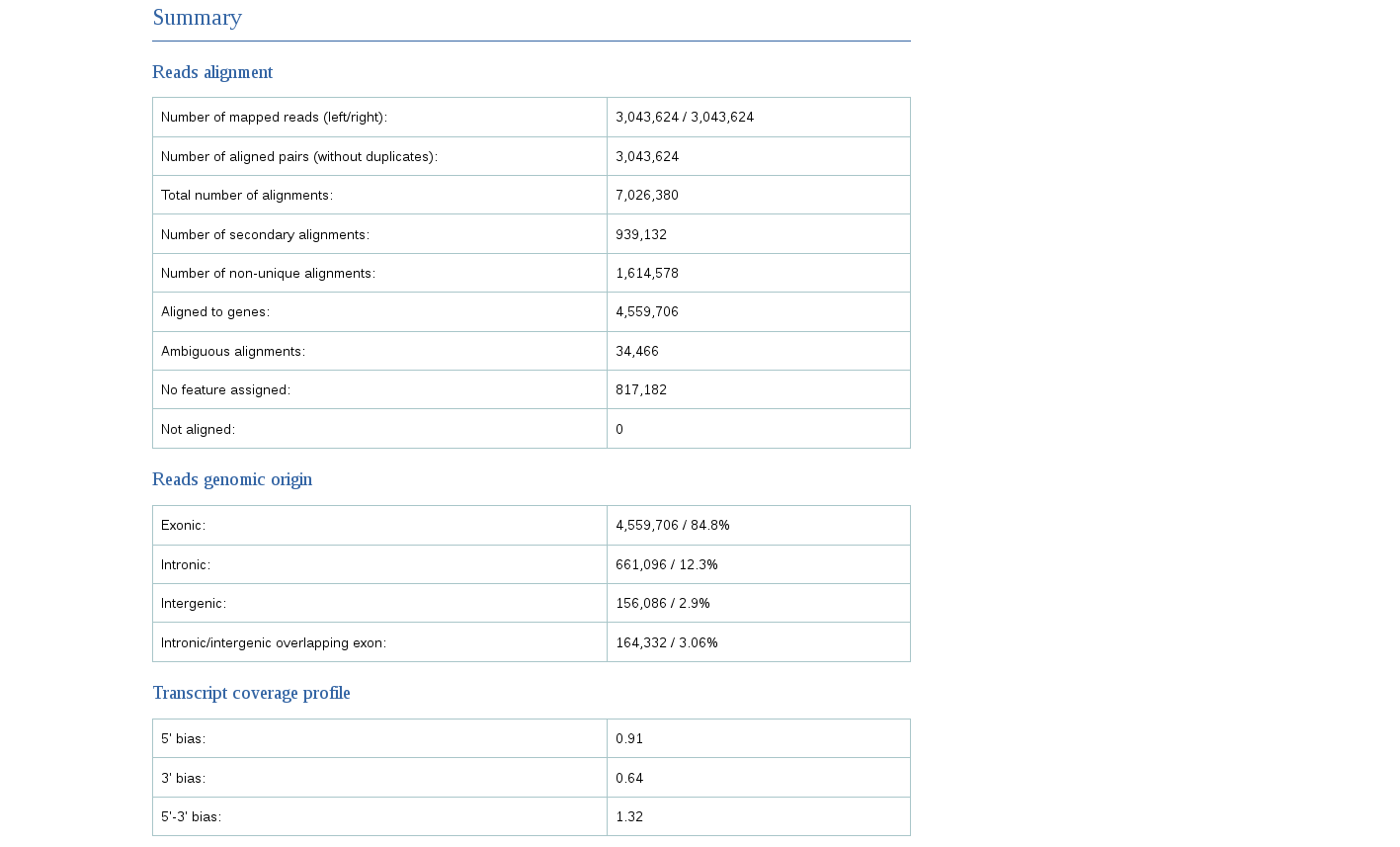
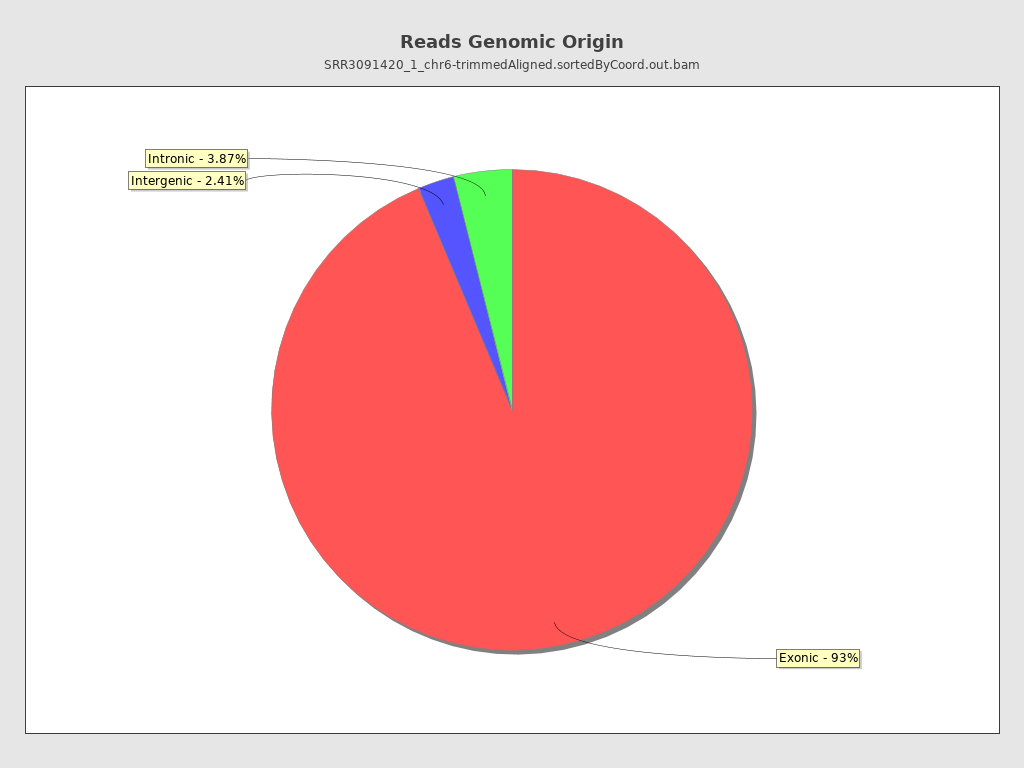
Finally, we can see that the majority of reads map to the exons.
IMPORTANT for running QualiMap on many samples (for detail, see QualiMap documentation
- Make sure to give to the output folder the name corresponding to a running sample; e.g., ./QC/SRR3091420_1_chr6; otherwise output files will be overwritten.
- If you run QualiMap in parallel for many samples, make sure to create a different tmp-folder for each sample; e.g., ./tmp/SRR3091420_1_chr6Aligned.
- QualiMap sorts BAM files by read names. To speed up this part of the program execution, you can use samtools to sort the BAM files in parallel and using multiple CPUs and then to give to QualiMap a BAM file sorted by read names and provide an option –sorted.

