9.1 Vectors
A vector is a sequence of data elements of the same type.
| 329 | 45 | 12 | 28 |
9.1.1 Creating a vector
- Values are assigned to a vector using the c function (c for combining elements). Open and close the round brackets, and place the different elements, separated by commas.
vector1 <- c(329, 45, 12, 28)You can create an empty vector with:
# empty vector
vecempty <- vector()
# empty numerical vector with 5 elements
vecnumempty <- vector(mode="numeric", length=5)
# empty character vector with 8 elements
veccharempty <- vector(mode="character", length=8)- Create a sequence of consecutive numbers:
vecnum <- 10:16
# shortcut for:
vecnum <- c(10, 11, 12, 13, 14, 15, 16)
# NOTES: both ends (10 and 16) are included- Character vectors: each element is entered between (single or double) quotes.
| mRNA | miRNA | snoRNA | lncRNA |
mynames <- c("mRNA", "miRNA", "snoRNA", "lncRNA")HANDS-ON
- Create a vector (choose the name) that contains a sequence of number, from 3 to 8, both included.
- Create vector myfruits as:
myfruits <- c(apple, pear, orange)What is happening? Why? Can you modify the line above so it doesn’t throw an error?
9.1.2 Vector manipulation
- A vector can be named: each element of the vector can be assigned a name (number or character)
# our previous numerical vector
vector1 <- c(329, 45, 12, 28)
# assign names
names(vector1) <- c("mRNA", "miRNA", "snoRNA", "lncRNA")
# use an object which already contains a vector
mynames <- c("mRNA", "miRNA", "snoRNA", "lncRNA")
names(vector1) <- mynames- Get the length (number of elements) of a vector
length(vector1)## [1] 4- Extracting elements from vector a
- extract elements using their position (index) in the vector:
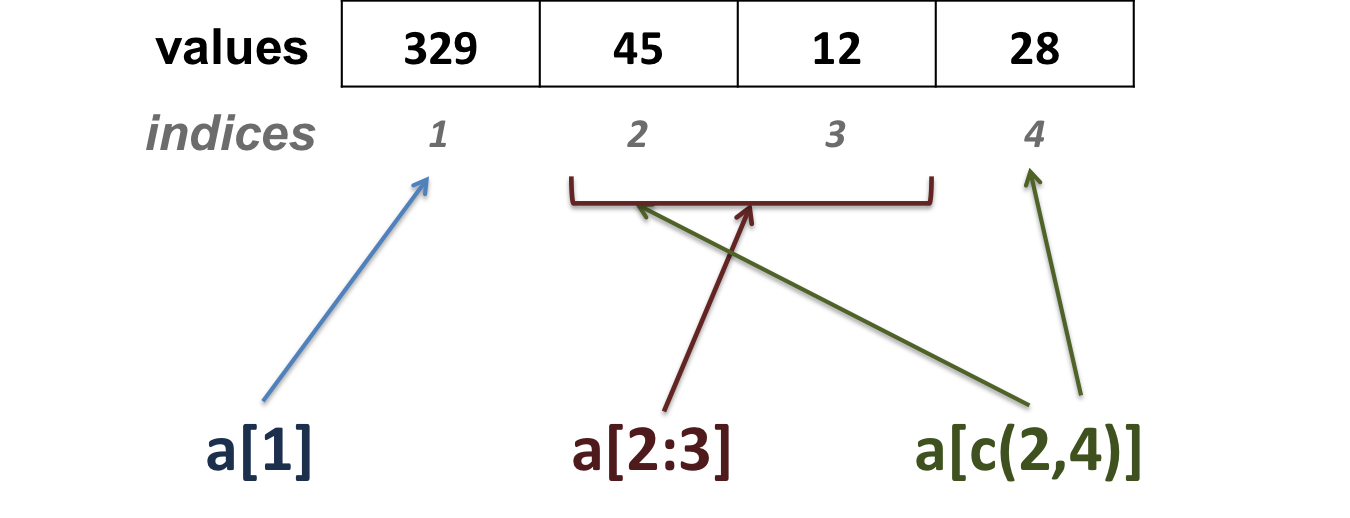
vector1[1]## mRNA ## 329vector1[c(1,3)]## mRNA snoRNA ## 329 12vector1[2:4]## miRNA snoRNA lncRNA ## 45 12 28- extract elements using their names:

vector1["mRNA"]## mRNA ## 329vector1[c("miRNA", "lncRNA")]## miRNA lncRNA ## 45 28 - extract elements using their position (index) in the vector:
- Reassigning a vector’s element
# here we re-assign the 2nd element of vector1 with 31.
vector1[2] <- 31
vector1["miRNA"] <- 31- Removing a vector’s element
# here we remove the 3rd element of vector1
# show in the console vector1 without its 3rd element:
vector1[-3]## mRNA miRNA lncRNA
## 329 31 28# reassign vector1 without its 3rd element
vector1 <- vector1[-3]- Show versus change
x[-2] ![]() x unchanged !
x unchanged !
x <- x[-2] ![]() x reassigned !
x reassigned !
9.1.3 Combining vectors
- You can combine 2 (or more) vectors into one. Here, we combine v1 and v2 to create a vector v3
v1 <- 2:5
v2 <- 4:6
v3 <- c(v1, v2)The elements of v2 are added after the elements of v1
- Likewise, you can add elements at the end of a vector
v3 <- c(v3, 19)9.1.4 Numeric vector manipulation
Logical operators
| Operator | Description |
|---|---|
| < | less than |
| <= | less than or equal to |
| > | greater than |
| >= | greater than or equal to |
| == | exactly equal to |
| != | not equal to |
| !x | not x |
| x | y | x OR y |
| x & y | x AND y |
- Which elements of a are equal to 2?
a <- 1:5
a == 2## [1] FALSE TRUE FALSE FALSE FALSE
- Which elements of a are superior to 2?
a <- 1:5
a > 2## [1] FALSE FALSE TRUE TRUE TRUE
- Extract elements of a vector that comply with a condition:
a <- 1:5
# extract the TRUE/FALSE vector
a >= 2## [1] FALSE TRUE TRUE TRUE TRUE# extract the actual sub-vector that complies the condition (TRUE values)
a[a >= 2]## [1] 2 3 4 5# count how many elements comply the condition
length(a[a >= 2])## [1] 4
HANDS-ON
Given the following vector containing a group of people’s ages:
ages <- c(18, 32, 12, 34, 42, 32, 17, 56, 76, 10)- How many people are over 18?
- How many people are under 60?
- How many people are between 25 and 45?
9.1.4.1 Operations on vectors
- Adding 2 to a vector adds 2 to each element of the vector:
a <- 1:5
a + 2## [1] 3 4 5 6 7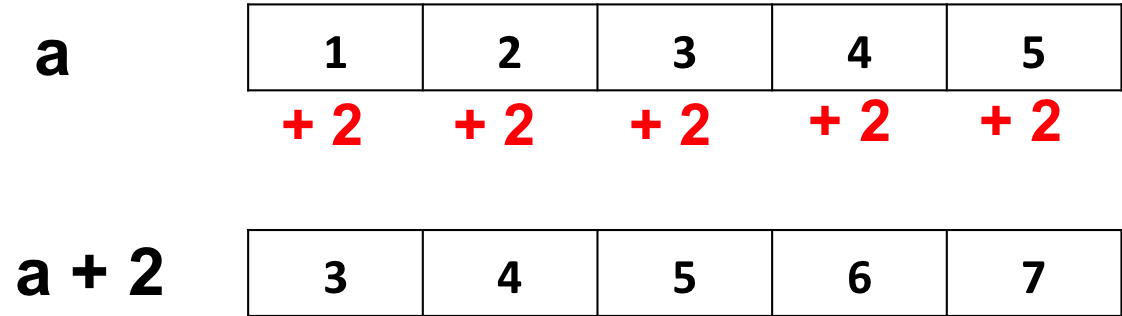
Same goes for subtractions, multiplications and divisions…
- Multiplying a vector by another vector of equal length
a <- c(2, 4, 6)
b <- c(2, 3, 0)
a * b## [1] 4 12 0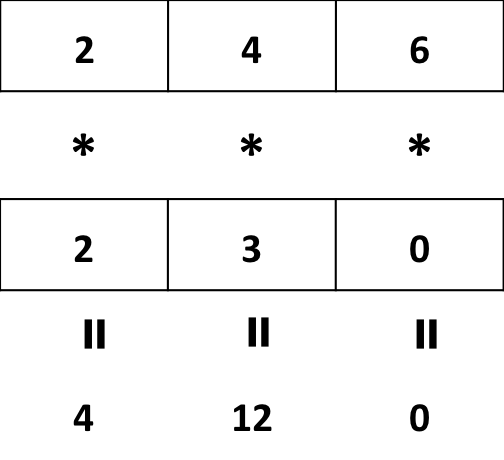
- Multiplying a vector by another shorter vector
a <- c(2, 4, 6, 3, 1)
b <- c(2, 3, 0)
a * b## Warning in a * b: longer object length is not a multiple of shorter object length## [1] 4 12 0 6 3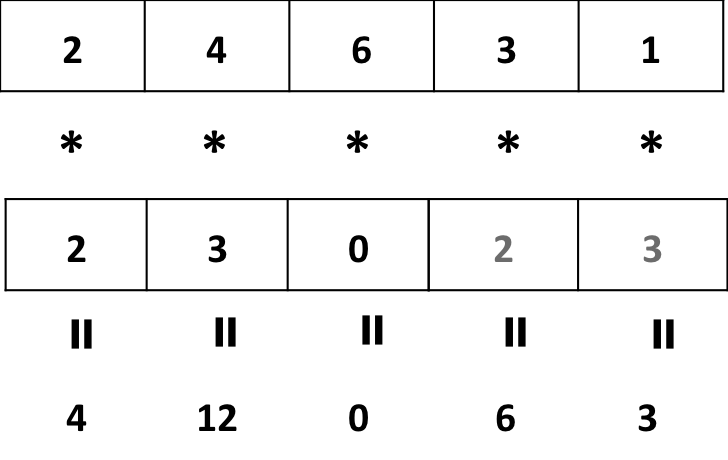
Vector a is “recycled” !
- Summary statistics
| Function | Description |
|---|---|
| mean(x) | mean / average |
| median(x) | median |
| min(x) | minimum |
| max(x) | maximum |
| var(x) | variance |
| summary(x) | mean, median, min, max, quartiles |
k <- c(1, 3, 12, 45, 3, 2)
summary(k)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 1.00 2.25 3.00 11.00 9.75 45.00HANDS-ON
Given our ages vector:
ages <- c(18, 32, 12, 34, 42, 32, 17, 56, 76, 10)- What is the average/mean age?
- What is percentage of variation? It is defined as (standard deviation / mean) * 100.
9.1.4.2 Comparing vectors
- The %in% operator
Which elements of a are also found in b ?
a <- 2:6
b <- 4:10
a %in% b## [1] FALSE FALSE TRUE TRUE TRUEYields a TRUE/FALSE vector of the size of a.
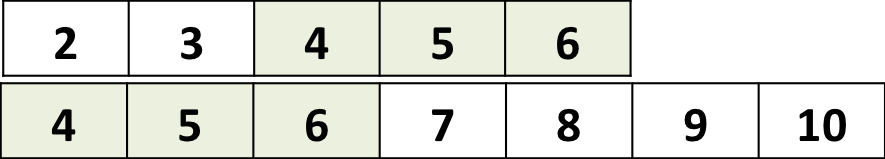
Retrieve actual elements of a that are found in b:
a <- 2:6
b <- 4:10
a[a %in% b]## [1] 4 5 6- Intersection:
Retrieve the intersection of 2 vectors:
intersect(a, b)## [1] 4 5 6- Check if 2 vectors are exactly identical
identical(a, b)## [1] FALSE9.1.5 Character vector manipulation
Character vectors are manipulated similarly to numeric ones.
- The %in% operator:
k <- c("mRNA", "miRNA", "snoRNA", "RNA", "lincRNA")
p <- c("mRNA","lincRNA", "tRNA", "miRNA")
k %in% p## [1] TRUE TRUE FALSE FALSE TRUEk[k %in% p]## [1] "mRNA" "miRNA" "lincRNA"- Select elements from vector m that are not exon
m <- c("exon", "intron", "exon")
m != "exon"## [1] FALSE TRUE FALSEm[m != "exon"]## [1] "intron"